
Presentations made painless
- Get Premium

115 Genetics Essay Topic Ideas & Examples
Inside This Article
Genetics is a fascinating and complex field of study that explores the inheritance and variation of traits in living organisms. From the discovery of DNA to the mapping of the human genome, genetics has revolutionized our understanding of how traits are passed down from one generation to the next.
If you are a student studying genetics, you may be tasked with writing an essay on a specific topic related to genetics. To help you get started, here are 115 genetics essay topic ideas and examples to inspire your writing:
- The impact of genetic engineering on agriculture
- The ethics of genetic manipulation in humans
- The role of genetics in determining intelligence
- Genetic disorders: causes and treatments
- The genetics of cancer
- The genetics of addiction
- Genetic testing in newborns: benefits and risks
- The genetics of aging
- The genetic basis of mental illness
- The role of genetics in personality traits
- The genetics of obesity
- The genetics of heart disease
- Genetic testing for hereditary diseases
- The genetics of skin color
- The genetics of eye color
- Genetic diversity in human populations
- The genetics of hair loss
- The genetics of height
- The genetics of blood type
- The genetics of taste preferences
- The genetics of athletic performance
- The genetics of hair texture
- The genetics of lactose intolerance
- The genetics of drug metabolism
- The genetics of alcoholism
- The genetics of diabetes
- The genetics of Alzheimer's disease
- The genetics of schizophrenia
- The genetics of bipolar disorder
- The genetics of autism
- The genetics of ADHD
- The genetics of dyslexia
- The genetics of Down syndrome
- The genetics of Turner syndrome
- The genetics of Klinefelter syndrome
- The genetics of cystic fibrosis
- The genetics of sickle cell anemia
- The genetics of hemophilia
- The genetics of Huntington's disease
- The genetics of Parkinson's disease
- The genetics of ALS
- The genetics of muscular dystrophy
- The genetics of color blindness
- The genetics of hemochromatosis
- The genetics of Marfan syndrome
- The genetics of Tay-Sachs disease
- The genetics of PKU
- The genetics of Angelman syndrome
- The genetics of Prader-Willi syndrome
- The genetics of Rett syndrome
- The genetics of Fragile X syndrome
- The genetics of Williams syndrome
- The genetics of Cri-du-chat syndrome
- The genetics of Patau syndrome
- The genetics of Edwards syndrome
- The genetics of Beckwith-Wiedemann syndrome
- The genetics of Prune Belly syndrome
- The genetics of Jacobs syndrome
- The genetics of Triple X syndrome
- The genetics of XYY syndrome
- The genetics of Wolf-Hirschhorn syndrome
- The genetics of Rubinstein-Taybi syndrome
- The genetics of Cornelia de Lange syndrome
- The genetics of Smith-Magenis syndrome
- The genetics of DiGeorge syndrome
- The genetics of Velocardiofacial syndrome
- The genetics of Duchenne muscular dystrophy
- The genetics of Becker muscular dystrophy
- The genetics of Myotonic dystrophy
- The genetics of Facioscapulohumeral muscular dystrophy
- The genetics of Oculopharyngeal muscular dystrophy
- The genetics of Limb-girdle muscular dystrophy
- The genetics of Emery-Dreifuss muscular dystrophy
- The genetics of Charcot-Marie-Tooth disease
- The genetics of Spinal muscular atrophy
- The genetics of Friedreich's ataxia
- The genetics of Ataxia telangiectasia
- The genetics of Niemann-Pick disease
- The genetics of Gaucher disease
- The genetics of Fabry disease
- The genetics of Pompe disease
- The genetics of Hurler syndrome
- The genetics of Hunter syndrome
- The genetics of Sanfilippo syndrome
- The genetics of Morquio syndrome
- The genetics of Maroteaux-L
Want to research companies faster?
Instantly access industry insights
Let PitchGrade do this for me
Leverage powerful AI research capabilities
We will create your text and designs for you. Sit back and relax while we do the work.
Explore More Content
- Privacy Policy
- Terms of Service
© 2024 Pitchgrade
Genetics - Free Essay Samples And Topic Ideas
Genetics is the study of genes, genetic variation, and heredity in organisms. Essays on genetics might delve into the fundamental principles of genetics, the discovery and function of DNA, or the development of genetic technologies like CRISPR. Other discussions could explore ethical issues related to genetic engineering, gene therapy, or genetic testing. Topics might also include the impact of genetics on medicine, agriculture, or understanding human evolution and diversity. The social implications of genetic research, the representation of genetics in popular culture, or the future of genetic science in addressing human health and environmental challenges could also be discussed. We have collected a large number of free essay examples about Genetics you can find at Papersowl. You can use our samples for inspiration to write your own essay, research paper, or just to explore a new topic for yourself.
Exploring the Intricacies of Genetics through DNA
Introduction The hereditary molecule that is tasked with carrying genetic instructions that are used in all living things in development, growth, reproduction and functioning is referred to as deoxyribonucleic acid (DNA). DNA molecules consist of two strands which are bipolar and are mostly coiled near to one another to form a spiral. This strands are referred to as polynucleotides simply because they are made of small units known as nucleotides. The information of the DNA is stored in this nucleotides. […]
The Physiology and Genetics Behind Alzheimer Disease
Alzheimer disease is a progressive and ultimately fatal brain disorder, in which communication between cells are halted and eventually lost. It is the most common form of dementia, and is generally (though not exclusively) diagnosed in patients over the age of 65. As communication amongst neurons is lost, symptoms such as inability to recall memories, make appropriate judgment, and proper motor function are lost and worsen over time. Affecting an estimated 2.4 million to 4.5 million Americans, with the number […]
GMO’s and World Hunger
As the world begins to feel the constraints of overpopulation and diminishing resources, the rate at which people are affected by chronic world hunger continues to grow exponentially (Geldof). Record climate change brought about by global warming and an increase in greenhouse emissions has increased the longevity of droughts, causing the desert to spread, and what small area of forest we have to left to soon run out (Gerry). According to research conducted at Harvard, the world population is estimated […]
We will write an essay sample crafted to your needs.
Connection between Genetics and Diabetes
Each single person has a specific set of genes; however, these genetics are greatly influenced by their families. Genetics can also be affected via one's environmental surroundings, as well. These genetics are associated with most diseases, such as cancer, kidney diseases, and psychologic diseases. Diabetes is no different. Genetics are not the only causative factor in diabetes, but it can alert healthcare members to look for this disease due to predisposition. According to the American Diabetes Association (2018), "Type 1 […]
Mitosis: Genetics Analysis & Principle
Introduction Mitosis is a process of nucleic division in animal or eukaryotic cells that occurs when a parent cell divides to produce two identical daughter cells. Without mitosis there wouldn't be a you or a me. Because during the cell division, mitosis, specifically separates the duplicated genetic material carried in the nucleus. While mitosis is taking place, there is no cell growth and all of the cellular energy is focused on cell division. The cell division processsd of mitosis is […]
The Mitosis Division
Introduction The mitosis division occurs in somatic cells and is opposed to the germ cell, which it undergoes Mitosis. Mitosis is following into the G2, and it occurs in the same time that cells begin to separate duplicate their content and divide them out. Toward the end of mitosis, the division of the cells yield identical diploid cells. (https://www.khanacademy.org/science/biology/cellular.../mitosis/a/phases-of-mitosis) .There is five stages that occur in mitosis, the first step is the Interphase. During the interphase, the cells is now […]
How Epigenetics May Affect Alzheimer’s Disease
Abstract Alzheimer's disease (AD) is a neurodegenerative disease affecting approximately 5.5 million people. Each year, more and more information is uncovered about AD and, recently, studies are attempting to validate the hypothesis that epigenetics significantly affects AD pathology. Recognizing the need for these studies the National Institute on Aging and Alzheimer's Association (NIA-AA) published a new research framework in an effort to redefine the disease based on biological marker, as opposed to syndromal markers. This review considers two published works, […]
Technology Evolution: Insights into Invisible Evolution and Epigenetics
From Divine Creation to Early Evolutionary Theories: Lamarck, Darwin, and the Quest for Understanding Until the eighteenth century, the general idea of how the world came to be was rooted in a Creator uniquely forming each type of species (Futuyma, 2017). The idea tended to be based on the Bible and the Christian faith. Many people believed a supreme being created the Earth and each different species, the species remaining unchanged throughout time. During the eighteenth century, several different scientists […]
What is Mitosis?
Mitosis is a complex division of a single cell, known as the ""mother"" cell, into two genetically similar cells, known as ""daughter"" cells. During this process the nuclear chromatin (located in the cell's nucleus and containing the DNA of the cell) is duplicated, and then split, creating 46 chromosomes(92 chromatids) for each of the ""daughter"" cells. The process of mitosis is made up of phases, sometimes including the preparation of the cell for division, interphase, while always including prophase, metaphase, […]
Genetics and Personality
In my initial position paper, I said that the past, present, and future is very important to personality. In addition, I theorized that it is important that we understand that the present is influenced by experience form the past; the present is influenced by one’s thought of the future; the concerns of the past and the future can be the result of one’s personality. Currently, I believe these theories are true but, they are not always accurate when predicting how […]
GMO Food Labeling
Genetically modified organisms, also known as GMO, are organisms that have been genetically altered to have a specific characteristic or trait. GMOs were first introduced in 1994 and no one knew about the potential health problems that could come. Nowadays more Americans worry about where their food comes from. Even though GMOs can help starvation and save labor costs, GMOs should be labeled because we don't know the long-term health effects, and GM foods can cause a numerous amount of […]
Pro GMO: Feeding the World
To fully understand the benefits GMO's we should first be able to define it. According to source, GMO's in reference to agriculture is, a plant and or microorganism whose genetic makeup has been modified in a laboratory using genetic engineering or transgenic technology. GMO's are not a newly introduced subject, in fact we have been eating GMO's for hundreds of years and we are still perfectly healthy. The public that is opposed to the use and of GMO crops, often […]
Social and Ethical Implications of GMO’s
There are biotechnology debates about genetically modified organisms in society and can be illustrated with the serious conflict between two groups that are voicing possible benefits and possible drawbacks to GMOs. First, are the Agricultural biotech companies that provide tools to farmers to yield bigger better crops but in the most cost-effective way, also known as Agri-biotech. Agri-biotech investors and their affiliated scientists versus the independent scientists, environmentalists, farmers, and consumers (Maghari 1). On one hand, you have the Agri-biotech […]
People with down Syndrome
This week, we learned a lot about genetics. But, there's always two sides to a story. There's the good side where the study of genetics can help us learn more about our past and our future. Then, there's the down side where we discover the shocking amount of diseases that are traveling among the human population. One down side in particular, Trisomy 21 or down syndrome, is a commonly heard disorder that results from the presence from either all or […]
DNA and Mutations
Occurrence of mutation. Mutation is the process that produces a gene or a chromosome set different from the wild type. For instance this allows us to measure the frequency of mutation occurance.a cell caring mutation can be used as probes to disassemble the constituent parts of a biological function and to examine their workings and interrelations.For a recessive mutation to give rise to a mutant phenotype in a diploid organism both alleles must carry the mutation but one copy of […]
Life with Down’s Syndrome
Worldwide, 100,000 babies are born with Down's Syndrome (DS), but it is rarely discussed or even acknowledged by those who do not have first-hand experience (Harvery, 2004, 43). Down's Syndrome was originally acknowledged by John Langdon Down in the 1800's, its causes were not discovered until 1959 by Jerome Lejeune, and its symptoms are continually being researched. You have come to this blog to educate yourself on how to best help your child with Down's Syndrome. Although Down Syndrome cannot […]
Dangerous Food GMO
Do you know that you eat often the GMO foods in everyday life. GMO was detected in our favorite Ramen and popular canola oil. What is GMO? It is made 'genetically modified foods' shorter and it is a genetically recombinant creature that manipulates the genes of common life into a new breed. According to this article, there is popular controversy now about the safety of GMO. On the affirmative, GMO foods are safe scientifically and provide food in starving nations. […]
Down Syndrome and the most Common Types
What is Down syndrome and what are the most common types? Down syndrome is a genetic disorder that is, a disorder arising from an abnormality in an individual's genetic material4. Human cells typically consist of 23 pairs of chromosomes. 1 chromosome in each pair comes from your father and the other comes from your mother, this results in the person having 3 copies of chromosome 21, instead of the usual 2 copies, in all cells. Some common physical traits of […]
Research Paper: Genetically Modified Organisms
Genetically modified organisms, otherwise referred to as GMOs, is a highly debated and researched topic throughout the world, however, highly prevalent in the United States today. It is plant, animals, or other organism in which their genetic makeup has been altered or modified by either genetic engineering or transgenic technology. GMOs are used either in the medical field or agriculturally, looking to cure diseases and create vaccines or attempt to get the healthiest or highest profit out a product. Prior […]
Chromosomal Abnormalities: down Syndrome
The human body is made up of trillions of cells. Cells are known as the basic building blocks of life. Every cell has a nucleus that contains genes, which store all of the genetic material (What is Down Syndrome, 2018). Genes are made up of deoxyribonucleic acid (DNA) that is packaged into chromosomes, which are responsible for inherited traits. Humans have 23 pairs of chromosomes, containing one chromosome from dad and one from mom, with a total of 46 altogether. […]
Genetically Modified Plants
Genetically Modified Organisms, better known as GMO's, are plants or animals whose gene code has been altered using genetic information from other living organisms such as bacteria, other plant species, animals, and even humans. Typically, genetic modification of plants involves the addition of genetic sequences coding for specific proteins that result in a desirable heritable trait. These proteins alter the biology of the plant to enhance characteristics that are beneficial to humans. But along with altered or added genes for […]
GMO’s: Feeding the World or Killing it
Many people today are often amazed by the amount of food and nutrients created a year for human consumption. The constant prominence of genetically modified (GMO) foods is not only intimidating, but confusing. The dictionary definition of GMO is genetically modified organism: an organism or microorganism whose genetic material has been altered by means of genetic engineering. Simply explained, foods are plants and animals that have had their genetic makeup artificially altered by scientists to make them grow faster, taste […]
Insulin-Dependent Diabetes Mellitus
Diabetes Mellitus 1, more specifically known as IDDM is a disorder concerning glucose homeostasis, which needs insulin therapy is generally seen in children. Diabetes is generally classified into 2 types IDDM (Insulin dependent diabetes mellitus) and the other NIDDM (Non-insulin dependent diabetes mellitus). Diabetes simply means an increase of glucose levels in the body as a result of the improper or no production of insulin from ones pancreatic ??-cells. The standard auto-immune response of type 1 diabetes is specific destruction […]
Mitosis and DNA Molecule
Replication is the copying of the genetic information from one DNA molecule into another DNA molecule. Mitosis and meiosis are similar in the fact that they make new cells. Mitosis replaces and repairs body cells, while meiosis makes gametes like eggs and sperm. Mitosis has an asexual reproduction, while meiosis has a sexual reproduction. These two reproductions have differences in its number of divisions, phases, chromosome numbers, etc. One difference between mitosis and meiosis would be their production of daughter […]
Environmental Science GMFS: our Savior or Destroyer
GMFs are genetically modified foods created by Herbert Boyer and Stanley Cohen back in 1973. This technological advance led to more genetically modified foods and organisms being created and manufactured. GMFs are created either by direct genetic code modification or selective breeding. Direct genetic code modification occurs when a certain part of the genetic code is cut out, copied into bacteria, made into bullets, loaded into a gene gun, and shot into a cell where the genetic information incorporates itself […]
Exome Sequencing to Identify Rare Mutations Associated with Breast Cancer Susceptibility
Abstract Background - Breast cancer predisposition has been known to be caused by hereditary factors. New techniques particularly exome sequencing have allowed/ helped us to identify new and novel variants that exhibit a phenotype. Method - In this review we discuss the advantages of exome sequencing and how it could help in understanding the familial breast cancer. In particular, we will discuss about the studies by Noh et al.(1), Thompson et al.(2), and Kiiski et al.(3), on how they have […]
The Tumor Suppressor Role of TAp73 in Two Types of Cancer
Transcription factor of p53 initiates apoptosis after receiving information about metabolic disorder or genetic damage, thus playing a critical role as tumor suppressor. p73 is a cousin of p53, shares lots of similarities with p53 including gene structure and amino acid level. Therefore, p73 is able to activate some p53 target genes by binding to p53-responsive elements when p53 is impaired. Also, p73 is rarely mutant compared to p53 in tumor cells. Whether p73 plays a role in tumor suppressor […]
The Potential of Chromosomal Therapy in down Syndrome
Down Syndrome (DS) is the most common chromosomal abnormality genetic disease in the world. In the United States roughly 6,000 babies are born with Down Syndrome, about 1 out of every 650-100 live births every year (Bittles et al. 2004). Older mothers are more likely to have a baby affected by the chromosomal disorder than younger mothers. In other words, the prevalence of Down Syndrome increases as the mother's age increases. The likelihood that a woman under 30 has a […]
“Born Gay” Michael Abrams
In the article “Born Gay” Michael Abrams proposes question why men become gay. Is this due to the gay gene/genes or due to the environment where they grew up or other biological traits? Is being a homosexual is nature or nurture? He was looking at several researches and projects to find the answer. The author states that becoming a gay is at least partially genetic. William Reiner explored how environment influences on sexuality by studying boys who were born with […]
Potential Mechanisms for Cancer Resistance in Elephants and Comparativage in Humans
It is expected that cancer risk would increase with body size and life span. Peto’s paradox describes the lack of correlation between body size, life span, and cancer risk (Caulin, 2011). The cellular mechanism behind this has only been experimentally demonstrated in rodents. TP53 is a gene that codes for the p53 protein. This gene is vital in tumor suppression, and is mutated in many human cancers (Jiang, 2018). Humans have one copy (2 alleles) of this gene. Both alleles […]
Related topic
Additional example essays.
- Compare And Contrast In WW1 And WW2
- Followership and Servant Leadership
- Why Abortion Should be Illegal
- Death Penalty Should be Abolished
- Research Paper #1 – The Trail of Tears
- A Class Divided
- The Significance of Following Orders in Daily Life and the Fire Service
- Ten Commandments of Computer Ethics: Steering Society to a Responsible Digital Future
- Self-awareness as the Main Factor of Emotional Intelligence
- Why Being on Time Is So Important
- "Just Walk on" by Brent Staples Summary: Racial Stereotypes and Their Impact
- 1984 Compared to Today
1. Tell Us Your Requirements
2. Pick your perfect writer
3. Get Your Paper and Pay
Hi! I'm Amy, your personal assistant!
Don't know where to start? Give me your paper requirements and I connect you to an academic expert.
short deadlines
100% Plagiarism-Free
Certified writers
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Published: 24 August 2020
The road ahead in genetics and genomics
- Amy L. McGuire 1 ,
- Stacey Gabriel 2 ,
- Sarah A. Tishkoff ORCID: orcid.org/0000-0002-1339-5959 3 , 4 ,
- Ambroise Wonkam ORCID: orcid.org/0000-0003-1420-9051 5 , 6 ,
- Aravinda Chakravarti ORCID: orcid.org/0000-0002-4264-2285 7 ,
- Eileen E. M. Furlong ORCID: orcid.org/0000-0002-9544-8339 8 ,
- Barbara Treutlein ORCID: orcid.org/0000-0002-3299-5597 9 ,
- Alexander Meissner ORCID: orcid.org/0000-0001-8646-7469 2 , 10 , 11 , 12 ,
- Howard Y. Chang ORCID: orcid.org/0000-0002-9459-4393 13 ,
- Núria López-Bigas ORCID: orcid.org/0000-0003-4925-8988 14 , 15 , 16 ,
- Eran Segal ORCID: orcid.org/0000-0002-6859-1164 17 &
- Jin-Soo Kim ORCID: orcid.org/0000-0003-4847-1306 18
Nature Reviews Genetics volume 21 , pages 581–596 ( 2020 ) Cite this article
83k Accesses
121 Citations
342 Altmetric
Metrics details
- Genetic techniques
In celebration of the 20th anniversary of Nature Reviews Genetics , we asked 12 leading researchers to reflect on the key challenges and opportunities faced by the field of genetics and genomics. Keeping their particular research area in mind, they take stock of the current state of play and emphasize the work that remains to be done over the next few years so that, ultimately, the benefits of genetic and genomic research can be felt by everyone.
The contributors
Amy L. McGuire is the Leon Jaworski Professor of Biomedical Ethics and Director of the Center for Medical Ethics and Health Policy at Baylor College of Medicine. She has received numerous teaching awards at Baylor College of Medicine, was recognized by the Texas Executive Women as a Woman on the Move in 2016 and was invited to give a TedMed talk titled “There is No Genome for the Human Spirit” in 2014. In 2020, she was elected as a Hastings Center Fellow. Her research focuses on ethical and policy issues related to emerging technologies, with a particular focus on genomic research, personalized medicine and the clinical integration of novel neurotechnologies.
Stacey Gabriel is the Senior Director of the Genomics Platform at the Broad Institute since 2012 and has led platform development, execution and operation since its founding. She is Chair of Institute Scientists and serves on the institute’s executive leadership team. She is widely recognized as a leader in genomic technology and project execution. She has led the Broad’s contributions to numerous flagship projects in human genetics, including the International HapMap Project, the 1000 Genomes Project, The Cancer Genome Atlas, the National Heart, Lung, and Blood Institute’s Exome Sequencing Project and the TOPMed programme. She is Principal Investigator of the Broad’s All of Us (AoU) Genomics Center and serves on the AoU Program Steering Committee.
Sarah A. Tishkoff is the David and Lyn Silfen University Associate Professor in Genetics and Biology at the University of Pennsylvania, Philadelphia, USA, and holds appointments in the School of Medicine and the School of Arts and Sciences. She is a member of the US National Academy of Sciences and a recipient of an NIH Pioneer Award, a David and Lucile Packard Career Award, a Burroughs/Wellcome Fund Career Award and an American Society of Human Genetics Curt Stern Award. Her work focuses on genomic variation in Africa, human evolutionary history, the genetic basis of adaptation and phenotypic variation in Africa, and the genetic basis of susceptibility to infectious disease in Africa.
Ambroise Wonkam is Professor of Medical Genetics, Director of GeneMAP (Genetic Medicine of African Populations Research Centre) and Deputy Dean Research in the Faculty of Health Sciences, University of Cape Town, South Africa. He has successfully led numerous NIH- and Wellcome Trust-funded projects over the past decade to investigate clinical variability in sickle cell disease, hearing impairment genetics and the return of individual findings in genetic research in Africa. He won the competitive Clinical Genetics Society International Award for 2014 from the British Society of Genetic Medicine. He is president of the African Society of Human Genetics.
Aravinda Chakravarti is Director of the Center for Human Genetics and Genomics, the Muriel G. and George W. Singer Professor of Neuroscience and Physiology, and Professor of Medicine at New York University School of Medicine. He is an elected member of the US National Academy of Sciences, the US National Academy of Medicine and the Indian National Science Academy. He has been a key participant in the Human Genome Project, the International HapMap Project and the 1000 Genomes Project. His research attempts to understand the molecular basis of multifactorial disease. He was awarded the 2013 William Allan Award by the American Society of Human Genetics and the 2018 Chen Award by the Human Genome Organization.
Eileen E. M. Furlong is Head of the Genome Biology Department at the European Molecular Biology Laboratory (EMBL) and a member of the EMBL Directorate. She is an elected member of the European Molecular Biology Organization (EMBO) and the Academia Europaea, and a European Research Council (ERC) advanced investigator. Her group dissects fundamental principles of how the genome is regulated and how it drives cell fate decisions during embryonic development, including how developmental enhancers are organized and function within the 3D nucleus. Her work combines genetics, (single-cell) genomics, imaging and computational approaches to understand these processes. Her research has advanced the development of genomic methods for use in complex multicellular organisms.
Barbara Treutlein is Associate Professor of Quantitative Developmental Biology in the Department of Biosystems Science and Engineering of ETH Zurich in Basel, Switzerland. Her group uses and develops single-cell genomics approaches in combination with stem cell-based 2D and 3D culture systems to study how human organs develop and regenerate and how cell fate is regulated. For her work, Barbara has received multiple awards, including the Friedmund Neumann Prize of the Schering Foundation, the Dr. Susan Lim Award for Outstanding Young Investigator of the International Society of Stem Cell Research and the EMBO Young Investigator Award.
Alexander Meissner is a scientific member of the Max Planck Society and currently Managing Director of the Max Planck Institute (MPI) for Molecular Genetics in Berlin, Germany. He heads the Department of Genome Regulation and is a visiting scientist in the Department of Stem Cell and Regenerative Biology at Harvard University. Before his move to the MPI, he was a tenured professor at Harvard University and a senior associate member of the Broad Institute, where he co-directed the epigenomics programme. In 2018, he was elected as an EMBO member. His laboratory uses genomic tools to study developmental and disease biology with a particular focus on epigenetic regulation.
Howard Y. Chang is the Virginia and D. K. Ludwig Professor of Cancer Genomics at Stanford University and an investigator at the Howard Hughes Medical Institute. He is a physician–scientist who has focused on deciphering the hidden information in the non-coding genome. His laboratory is best known for studies of long non-coding RNAs in gene regulation and development of new epigenomic technologies. He is an elected member of the US National Academy of Sciences, the US National Academy of Medicine, and the American Academy of Arts and Sciences.
Núria López-Bigas is ICREA research Professor at the Institute for Research in Biomedicine and Associate Professor at the University Pompeu Fabra. She obtained an ERC Consolidator Grant in 2015 and was elected as an EMBO member in 2016. Her work has been recognized with the prestigious Banc de Sabadell Award for Research in Biomedicine, the Catalan National Award for Young Research Talent and the Career Development Award from the Human Frontier Science Program. Her research focuses on the identification of cancer driver mutations, genes and pathways across tumour types and in understanding the mutational processes that lead to the accumulation of mutations in cancer cells.
Eran Segal is Professor in the Department of Computer Science and Applied Mathematics at the Weizmann Institute of Science, heading a multidisciplinary laboratory with extensive experience in machine learning, computational biology and analysis of heterogeneous high-throughput genomic data. His research focuses on the microbiome, nutrition and genetics, and their effect on health and disease and aims to develop personalized medicine based on big data from human cohorts. He has published more than 150 publications and received several awards and honours for his work, including the Overton and the Michael Bruno awards. He was recently elected as an EMBO member and as a member of the Israel Young Academy.
Jin-Soo Kim is Director of the Center for Genome Engineering in the Institute for Basic Science in Daejon, South Korea. He has received numerous awards, including the 2017 Asan Award in Medicine, the 2017 Yumin Award in Science and the 2019 Research Excellence Award (Federation of Asian and Oceanian Biochemists and Molecular Biologists). He was featured as one of ten Science Stars of East Asia in Nature ( 558 , 502–510 (2018)) and has been recognized as a highly cited researcher by Clarivate Analytics since 2018. His work focuses on developing tools for genome editing in biomedical research.
Similar content being viewed by others

A brief history of human disease genetics
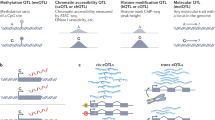
Molecular quantitative trait loci
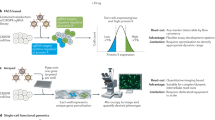
A new era in functional genomics screens
Making genomics truly equitable.
Amy McGuire. For the field of genetics and genomics, the first decade of the twenty-first century was a time of rapid discovery, transformative technological development and plummeting costs. We moved from mapping the human genome, an international endeavour that took more than a decade and cost billions of dollars, to sequencing individual genomes for a mere fraction of the cost in a relatively short time.
During the subsequent decade, the field turned towards making sense of the vast amount of genomic information being generated and situating it in the context of one’s environment, lifestyle and other non-genetic factors. Much of the hype that characterized the previous decade was tempered as we were reminded of the exquisite complexity of human biology. A vision of medicine driven by genetically determined risk predictions was replaced with a vision of precision in which genetics, environment and lifestyle all converge to deliver the right treatment to the right patient at the right time 1 .
As we embark on the third decade of this century, we are now faced with the prospect of being able not only to more accurately predict disease risk and tailor existing treatments on the basis of genetic and non-genetic factors but also to potentially cure or even eliminate some diseases entirely with gene-editing technologies.
These advancements raise many ethical and policy issues, including concerns about privacy and discrimination, the right of access to research findings and direct-to-consumer genetic testing, and informed consent. Significant investment has been made to better understand the risks and benefits of clinical genomic testing, and there has been vigorous debate about the ethics of human gene editing, with many prominent scientists and bioethicists calling for a moratorium on human germline editing until it is proven to be safe and effective and there is broad societal consensus on its appropriate application 2 .
These are all important issues that we need to continue to explore, but as the technologies that have been developed and tested at warp speed over the past two decades begin to be integrated into routine clinical care, it is imperative that we also confront one of the most difficult and fundamental challenges in genomics, in medicine and in society — rectifying structural inequities and addressing factors that privilege some while disadvantaging others. The genomics of the future must be a genomics for all, regardless of ethnicity, geography or ability to pay.
This audacious goal of making genomics truly equitable requires multifaceted solutions. The disproportionate burden of illness and death among racial and ethnic minorities associated with the global COVID-19 pandemic 3 and recent protests against police brutality towards African American citizens 4 have strengthened the antiracism movement and amplified demands for racial equity.
To be part of this movement and effect change will require humility. We must actively listen and learn from each other, especially when it is uncomfortable and our own complicity may be implicated. It will require solidarity and a recognition that we are all connected through our common humanity. And it will require courage. It may seem like a platitude, but it is true that nothing will change unless actual change is made. If we continue to do things as they have always been done, we will end up where we have always been. It is time to step into the discomfort and dare to do something different.
So what can we do differently to make genomics more equitable? I propose three areas where we should focus attention to address this important question. First, we must ensure equitable representation in genomic research. Examining 2,511 studies involving nearly 35 million samples from the GWAS Catalog in 2016, Popejoy and Fullerton found that the vast majority (81%) come from individuals of European descent, with only 5% coming from non-Asian minority populations 5 . This has created an ‘information disparity’ that has an impact on the reliability of clinical genomic interpretation for under-represented minorities 6 . The US National Institutes of Health (NIH) has invested in efforts to increase diversity in genomic research, but to be successful these efforts must be accompanied by serious attention to earning the trust of disadvantaged and historically mistreated populations. This will require, at a minimum, more meaningful engagement, improved transparency, robust systems of accountability, and a commitment to creating opportunities that promote and support a genomics workforce that includes scientists and clinicians from under-represented populations.
It is insufficient to achieve diverse representation in genomic research; however, there must also be equitable access to the fruits of that research. An analysis of the US Centers for Disease Control and Prevention’s 2018 Behavioural Risk Factor Surveillance System found that non-elderly adults from self-identified racial or ethnic minority groups are significantly less likely to see a doctor because of cost than non-elderly white adults 7 . This finding reflects how the structure and financing of health care in the United States perpetuates inequities and contributes to the larger web of social injustice that is at the heart of the problem. Even when socio-economic factors are controlled for, racial disparities in access to genetic services persist 8 . Large-scale, sustained research is needed to better understand and actively address the multitude of factors that contribute to this, including issues related to structural racism, mistrust, implicit and explicit bias, a lack of knowledge of genetic testing, and concerns about misuse of genetic information.
Finally, and perhaps most daunting, we must strive to achieve more equitable outcomes from genomic medicine. Many racial and ethnic minorities disproportionately experience chronic disease and premature death compared with white individuals. Disparities also exist by gender, sexual orientation, age, disability status, socio-economic status and geographical location. Health outcomes are heavily influenced by social, economic and environmental factors. Thus, although providing more equitable access to genomic services and ensuring more equitable representation in genomic research are necessary first steps, they are not enough 9 . Genomics can only be part of the solution if it is integrated with broader social, economic and political efforts aimed at addressing disparities in health outcomes. For genomics to be truly equitable, it must operate within a just health-care system and a just society.
we must strive to achieve more equitable outcomes from genomic medicine
Genome sequencing at population scale
Stacey Gabriel. Twenty years ago, I finished a PhD project that involved laboriously sequencing one gene — a rather complicated one, RET — in a couple of hundred people to catalogue pathogenic variants for Hirschsprung disease. This work required designing primers on the basis of genome sequence data as they were gradually released, amplifying the gene exon by exon (all 20!), running sequencing gels and manually scoring sequence changes. The notion of sequencing the whole genome to catalogue sequence changes was something to wish for in our wildest dreams.
Thanks to great strides in technology and the hard work of geneticists, engineers, epidemiologists and clinicians, much progress has been made; huge numbers of genomes (and exomes) have been sequenced across the world. Disease gene-finding projects such as my graduate work are now done routinely, rather than one gene at a time, using whole-exome or whole-genome sequencing (WGS) in families and affected individuals, enabling the identification of genes and causative mutations in thousands of Mendelian diseases and some complex diseases.
But the real promise of genome sequencing lies in true population-scale sequencing, ultimately at the scale of tens of millions of individuals, whereby genome sequencing of unselected people enables the unbiased, comprehensive study of our genome and the variation therein. It provides a ‘lookup table’ to catalogue disease-causing and benign variants (our ‘allelic series’). The genome sequence should become part of the electronic health record; it is a stable, persistent source of information about a person akin to physical measurements such as weight or blood pressure, exposures such as smoking or alcohol use, and (in many ways better than) self-reported family history.
the real promise of genome sequencing lies in true population-scale sequencing, ultimately at the scale of tens of millions of individuals
What can we learn? What needs to be solved? Even fairly small numbers of genomes aggregated in a consistent and searchable form have enabled a new way to use and interpret genomic data, just in the past couple of years providing a glimpse at the future. Efforts such as gnomAD 10 are a start — this database contains data from more than 15,000 genomes and 125,0000 exomes. With this resource, the frequency of genetic variants within populations is readily available. A clinician interpreting the genome of a patient can ask whether a variant has been observed before. The data provide a starting point for assessing the functional impact of classes of genetic variation and the ability to ask questions about ‘missing’ genetic variation where there is constraint.
Coupled with clinical data, building up population-scale databases of genomic plus clinical information will fuel the application of better risk interpretation using polygenic risk scores (PRSs) 11 . More routine WGS will shorten the ‘diagnostic odyssey’, in which patients suffer through rounds of testing and parents are left uncertain about future reproductive planning. More efficient clinical trials might be built using genomic information. With existing genomic information on all individuals in a health system, trials could be designed in a way that selects individuals most likely to have an event. This enrichment could provide more promising, shorter, smaller and cheaper trial design.
These databases must also rapidly be built in such a way that is representative of the population, representing the actual racial and ethnic diversity, not just what was available as banked sample collections. These are well known to be predominantly European-descent samples and thus preclude application of risk prediction tools in non-white individuals and have limited the ability to find population-specific genetic associations, such as those that have been demonstrated in type 2 diabetes mellitus (T2DM) 12 .
We have to solve important issues — data sharing, privacy and getting the data to scale. Sharing genomic and clinical data is of key importance to drive forward discovery and our understanding of how to use these data in the health-care setting. To do this well and responsibly, trust must be built and maintained through adherence to the rights of privacy, protection and non-discrimination. Progress is being made through the creation of data platforms and the development of frameworks for data protection and sharing; for example, by the work of the Global Alliance for Genomics and Health (GA4GH).
Several large biobanks are already being established to launch population-scale efforts. The UK Biobank is a vanguard programme that contains genotype data, questionnaire-based health and physical measurements on 500,000 individuals and some linkage to their medical records. Other efforts such as the All of Us research programme have been launched with goals directed at true population-based representation, and biobanks that link genomic data to comprehensive medical records in specific health-care systems (for example, Geisinger) or in specific countries or regions (for example, Estonia and Iceland) are also under way.
A big piece of this puzzle is generating comprehensive genome sequence data in these programmes and far beyond. For this aim, large-scale, affordable sequencing is key. No problem, right? Is sequencing not always getting cheaper? The problem is that this assumption is no longer true. We have got to where we are today because for a long time, from 2008 to 2013, sequencing costs dropped exponentially. However, in recent years, the sequencing cost curve has flattened, as is apparent in publicly reported cost estimates provided by the US National Human Genome Research Institute 13 . The cost per megabase of sequence data has remained largely unchanged since around 2016, hovering around a list price of US$0.01 per megabase, which translates to a US$1,000 genome. Gone are the days of our field touting the impressive decrease of cost in comparison with Moore’s law, and this development is worrying.
Some discounting does happen at considerable volume, and whole genomes can be priced in the range of US$500 to US$700. However, large projects (more than 500,000 samples) sequenced at these prices are few and far between, and are generally dependent on pharmaceutical or biotech funding, which can bring with it restrictions on data sharing. It is my belief that a fivefold to sevenfold reduction in total costs is needed to unlock more sequencing at the population scale and, ultimately, for genome sequencing to be more widely applied in the health-care setting. At US$100 per genome, the cost represents less than 1% of the annual average health-care expenditure per person in the United States, and a genome sequence is a one-time investment that can be referenced again and again over the entire lifespan of a person. Getting that cost curve down will be important to inspire health-care systems to adopt genome sequencing routinely.
I see three main drivers that will get us to US$100 per genome: innovation, scale and competition.
Innovation . Generating sequence data requires multiple components, and there are multiple areas ripe for innovation. Sample preparation can be improved through more efficient methods that decrease the labour required, or miniaturization can decrease the cost of the reagents used in library preparation. Developments to decrease data processing costs are also ripe for innovation. Recently, we showed that processing using optimized computing power lowered the time and cost of creating a sequence file by ~50% (S.G., unpublished observations). While decreases in the costs of sample preparation and data processing are important, they represent a small component of the total cost. Roughly 70% of the cost of sequencing a human genome is the sequencing reagent (flow cell) and the instrument. Appreciable cost decrease is made possible only by decreasing these marginal costs, as was demonstrated in the period from 2010 to 2014, when flow-cell densities doubled and sequencing cost dropped by an order of magnitude (US$100 per gigabase to US$10 per gigabase).
Scale . One component of cost is the fixed cost borne by the sequencing centre or the sequencing vendor. With high scale, centres can become more efficient and offset costs such as the costs of personnel, equipment and facilities. Scale can also result in volume discounting of the reagents, although this process is tightly controlled and approached cautiously depending on overall market dynamics.
Competition . Innovation and scale can only achieve so much. The cost of generating the data (the cost per gigabase) dominates and thus must come down considerably. The current market requires alternative options to drive this advance. Presently, the market for short-read sequencing is lacking viable, proven competition that would force flow-cell densities and machine yield to be increased and put pressures on volume discounting. While options for long-read sequencing exist and play a role in particular applications, such as de novo sequencing and structural variant resolution, they are at present far from cost competitive and, therefore, do not apply pressure to bring down the cost of routine WGS.
We need innovation, great economies of scale and/or real competition to come to play in the marketplace. When it comes to sequencing technology, particularly at a large scale, we cannot be complacent and work around the current barriers to realize small gains and one-off wins. This might involve specific types of investment beyond just financial ones; adopting and vetting new technology requires time, creativity, commitment and patience. It is a challenge for our community to take on now. In 5 years’ time, I hope we can look back at the era of the US$100 genome and progress towards real population-scale databases that fuel discovery, enriching our knowledge of the human allelic series and, importantly, the routine use of genomic data in the health-care setting.
A global view of human evolution
Sarah Tishkoff. The past 10 years saw an exponential increase in SNP array and high-coverage WGS data owing to innovations in genomic technologies. It is now possible to generate WGS data from tens of thousands of individuals (for example, GenomeAsia 100K 14 and NIH TOPMed 15 ). An increase in medical biobanks with access to electronic health records (for example, the UK Biobank 16 , the Million Veteran Project 17 and BioBank Japan 18 ) is enabling the mapping of hundreds of genetic associations with complex traits and diseases, as well as phenome-wide association studies 19 to map pleiotropic associations of phenotypes with genes. The genetic associations identified in these and other studies have been used to calculate PRSs for predicting complex phenotypes and risk of diseases.
Yet despite these advances, as of 2019, nearly 80% of individuals in genome-wide association studies (GWAS) were of European ancestries, ~10% were of East Asian ancestries, ~2% were of African ancestries, ~1.5% were of Hispanic ancestries and less than 1% were of other ancestries 20 . There is also a strong European bias in genomic reference databases, such as gnomAD and GTEx . These biases limit our knowledge of genetic risk factors for disease in ethnically diverse populations and could exacerbate health inequities 20 . Furthermore, PRSs that were estimated using European data do not accurately predict phenotypes and disease risk in non-European populations, performing worst in individuals with African ancestry 21 . The lack of transportability of PRSs across ethnic groups is likely due to differences in patterns of linkage disequilibrium and haplotype structure (resulting in different SNPs tagging causal variants), differences in allele frequencies, gene × gene effects and gene × environment effects. It is also possible that the genetic architecture of complex traits and diseases may differ across ethnic groups owing to different demographic histories and adaptation to diverse environments.
Although there have been initiatives to increase inclusion of ethnically diverse populations in human genomics research (for example, the NIH TOPMed 15 and H3Africa consortia), Indigenous populations remain under-represented. Great care must be taken to ensure that genomic research of minority and Indigenous populations is conducted in an ethical manner. This involves establishing partnerships with local research scientists, being sensitive to local customs and cultural concerns, obtaining both community and individual consent, and returning results to communities that participated when possible. In addition, there should be training and capacity building so that genomic research can be conducted locally, where feasible.
A particular area of focus in the future should be developing tools and resources that make genomic data and analyses accessible in low- and middle-income countries. We have to ensure that all people benefit from the genomics revolution and advances in precision medicine and gene editing. Thus, several of the biggest challenges in the next decade will be (1) to increase inclusion of ethnically diverse populations in human genomics research; (2) the generation of more diverse reference genomes using methods that generate long sequencing reads, and haplotype phasing, to account for the large amount of structural variation that likely exists within and between populations; (3) the training of a more diverse community of genomic research scientists; and (4) the development of better methods for accurately predicting phenotypes and genetic risk across ethnically diverse populations and for distinguishing gene × environment effects.
The inclusion of ethnically diverse populations, including Indigenous populations, is also critical for reconstructing human evolutionary history and understanding the genetic basis of adaptation to diverse environments and diets. While there have been a number of success stories for identifying genes of large effect that play a role in local adaptation (for example, lactose tolerance and sickle cell disease (SCD) associated with malaria resistance), identifying signatures of polygenic selection has been considerably more challenging 22 . Genomic signatures of polygenic adaptation are based on the ability to detect subtle shifts in allele frequencies at hundreds or thousands of loci with minor effect on the phenotype of a complex trait and to determine whether that shift is a result of demography or natural selection. A more daunting challenge arises from the same issues of portability of PRSs described earlier — variants associated with a complex trait may not tag well across ethnic groups and/or the genetic architecture of a trait may differ in different populations. Furthermore, it has recently been shown that uncorrected population stratification can result in a false signal of polygenic selection 23 . For example, several studies have identified signatures of polygenic adaptation for height across European populations (selection for increased height in northern Europeans and for decreased height in southern Europeans). However, it was recently shown that these results were influenced by population structure that could not be easily corrected using standard approaches, particularly for SNPs below genome-wide levels of significance 23 . When this analysis was repeated with variants identified in a more homogenous set of individuals of European ancestry from the UK Biobank, these signatures of polygenic adaptation were erased 23 . Thus, methods for detecting polygenic adaptation that are less biased by population structure and by population ascertainment bias will need to be developed in the future. These studies will also benefit from inclusion of more ethnically diverse populations in GWAS and identification of better tag SNPs as described earlier. A challenge of inclusion of minority populations in GWAS is that sample sizes are often small relative to majority populations. However, the high levels of genetic diversity and extremes of phenotypic diversity observed in some populations, particularly those from Africa, make them particularly informative for GWAS. For example, a GWAS of skin pigmentation in fewer than 1,600 Africans was informative for identifying novel genetic variants that affect skin colour, including a previously uncharacterized gene, MFSD12 (ref. 24 ). Thus, genomic studies in the future must make inclusion of minority populations a priority.
A challenge in both GWAS and selection scans has been the identification of causal genetic variants that directly have an impact on variable traits. Most of these variants are in non-coding regions of the genome. The development of high-throughput approaches, such as massively parallel luciferase expression assays to identify gene regulatory regions and high-throughput CRISPR screens in vitro and in vivo to identify functional variants influencing the trait of interest, will be useful 25 . There is also a need to better understand cell type-specific variation and gene regulation at the single-cell level, including response to stimuli such as immune, pharmacological and nutrient challenges, in ethnically diverse populations. However, these approaches are still limited by the need to have informative cell lines. This can be particularly challenging to obtain for Indigenous populations living in remote regions. Improvements in the differentiation of induced pluripotent stem cells (iPS cells) into assorted cell types and into organoids will be important for facilitating functional genomic studies. Establishment of iPS cells and organoids from diverse non-human primate species will also be informative for comparative genomic studies to identify the evolution of human-specific traits such as brain development and cognition. However, iPS cell-derived cells may not accurately reflect the impact of mutations acting on developmental phenotypes, which will require development of more efficient in vivo approaches in model organisms.
Perhaps the biggest revolution in the study of recent human evolutionary history has been the development of methods that make it feasible to sequence and/or obtain targeted genotypes from ancient DNA samples. The generation of high-coverage reference genomes for archaic hominid species such as Neanderthals and Denisovans, located in Eurasia, has made it feasible to identify archaic introgressed segments within the genomes of non-Africans. Some of these regions have been shown to play a role in adaptive traits such as adaptation to high altitude and immune response 26 . Furthermore, there has been an explosion of studies of ancient genetic variation in Europeans within the past 30,000 years that has demonstrated a much more complex model of the peopling of Europe, and the recent evolution of adaptive traits, than previously known from the archaeological record or from studies of modern populations 27 . The biggest challenge has been the inability to get high-quality ancient DNA from regions with a tropical climate, such as Africa and Asia. While there has been success in analysing DNA samples as old as 15,000 years in Africa, which has been informative for tracing recent migration and admixture events 28 , the lack of a more ancient African reference genome makes it very challenging to detect archaic introgression, which currently relies on statistical modelling approaches. Thus, the biggest challenge in the next 10 years will be the successful sequencing of ancient DNA more than 20,000 years old from all regions of the world, so that we may have a better understanding of the complex web of population histories from across the globe.
African genomics — the next frontier
Ambroise Wonkam. To fully meet the potential of global genetic medicine, research into African genomic variation is a scientific imperative, with equitable access being a major challenge to be addressed. Studying African genomic variation represents the next frontier of genetic medicine for three major reasons: ancestry, ecology and equity.
On the basis of a ‘pan-genome’ generated from 910 individuals of African descent, at least 300 million DNA variants (10%) are not found in the current human reference genome 29 , and 2–19% of the genome of ancestral Africans derives from poorly investigated archaic populations that diverged before the split of Neanderthals and modern humans 30 . Neanderthal genome contributions make up ~2% of the genome in present-day Europeans and are enriched for variations in genes involved in dermatological phenotypes, neuropsychiatric disorders and immunological functions 31 . Once technical challenges in sequencing poor-quality DNA have been overcome and approaches to investigate the genomic contribution of African archaic populations have been refined, it is likely that associations between variants in ancient African DNA and human traits or diseases will be found, providing insights that can benefit modern-day humans.
As a consequence of the 300,000–500,000 years of genomic history of modern humans in Africa, ancestral African populations are the most genetically diverse in the world. By contrast, there is an extreme genetic bottleneck, resulting in much less variation, in all non-African populations who evolved from the thousands of humans who migrated out of Africa approximately 70,000 years ago. Current PRSs, which aim to predict the risk for an individual of a specific disease on the basis of the genetic variants that individual harbours, exhibit a bias regarding usability and transferability across populations, as most PRSs do not account for multiple alleles that are either limited or of high frequency among Africans. A GWAS on the genetic susceptibility to T2DM identified a previously unreported African-specific significant locus, while showing transferability of 32 established T2DM loci 32 . In addition, nonsense mutations found commonly among Africans in PCSK9 , which are rare in Europeans 33 , are associated with a 40% reduction in plasma levels of low-density lipoprotein, supporting PCSK9 as a target for dyslipidaemia therapeutics. In the largest GWAS meta-analysis for 34 complex traits, conducted in 14,345 Africans, several loci had limited transferability among cohorts 34 , further illustrating that genomic variation is highest among Africans compared with other populations. As a consequence, linkage disequilibrium is lower in Africans, which improves fine mapping and identification of causative variants. Indeed, while only 2.4% of participants in large GWAS are African individuals, they account for 7% of all associations 35 . Moreover, whole-exome sequencing of nearly 1,000 African study participants of Xhosa ancestry with schizophrenia found very rare damaging mutations in multiple genes 36 , a finding that could be replicated in a Swedish cohort of 5,000 individuals. In comparison, results for the Xhosa cohort yielded larger effect sizes, which shows that for the same number of cases and controls, the greater genetic variation in African populations provides more power to detect genotype–phenotype relationships. Therefore, millions of African genomes must be sequenced, with genotyping and analysis tools optimized for their interrogation.
Greater availability of African genomes will improve our understanding of genomic variation and complex trait associations in all populations but will also support research into common monogenic diseases. The discovery of a single African origin of the SCD mutation, about 5,000–7,000 years ago, not only suggested recent migration and admixture events between Africans and Mediterranean and/or Middle Eastern populations but also enhanced our understanding of genetic variation in general as well as its potential impact on haemoglobinopathies 37 . For example, variants in the HBB -like gene cluster linked with high levels of fetal haemoglobin have been associated with less severe SCD; because the level of fetal haemoglobin is under genetic control, it is amenable to therapeutic manipulation by gene editing 38 . Moreover, knowledge of an individual’s genetic variants can have an impact on secondary prevention of and treatment strategies for SCD. For example, variants in APOL1 and HMOX1 and co-inheritance of α-thalassaemia are associated with kidney dysfunctions 39 ; stroke in SCD is associated with targeted genetic variants used in a Bayesian model; and overall SCD mortality has been associated with circulating transcriptomic profiles. It is estimated that 75% of the 305,800 babies with SCD born each year are born in Africa; SCD in Africa will serve as a model for understanding the impact of genetic variation on common monogenic traits and help to illustrate the multiple layers of genomic medicine implementation.
Greater availability of African genomes will improve our understanding of genomic variation and complex trait associations in all populations
Exploring African genomic diversity will also increase discovery of novel variants and genes for rare monogenic conditions. Indeed, allelic and locus heterogeneity display important differences in African individuals compared with other populations; for example, mutations in GJB2 account for nearly 50% of cases of congenital non-syndromic hearing impairment among Eurasians but are nearly non-existent in Africans, and there is evidence that novel variants in hearing impairment-associated genes are more likely to be found in Africans than in populations of European or Asian ancestries 40 . Higher fertility rate, consanguinity practices and regional genetic bottlenecks will improve novel gene discovery for monogenic diseases in Africa, as well as disease–gene pair curation, and will address existing challenges surrounding database biases and inference of variant deleteriousness, which have led to the misclassification of variants.
Differential population genomic variant frequencies are shaped by natural evolutionary selection as an adaptation to environmental pressures. The African continent follows a North–South axis, which is associated with variable climates and biodiversity, both motors of natural selection. This specific African ecology has shaped genetic variation accordingly, which can have a detrimental or positive impact on health. Obvious examples are variants that cause SCD but confer resistance to malaria 37 , APOL1 variants that are protective against trypanosomes (the parasites that cause sleeping sickness) 41 and variants of OSBPL10 and RXRA that protect against dengue fever 42 . Unfortunately, APOL1 variants also increase susceptibility to chronic kidney disease in populations of African ancestry 39 , 41 . A better understanding of the functional impact of genetic variants specific to African populations, particularly those that have been selected under environmental pressure, and the way they interact with each other is needed and will have a positive impact on genetic medicine practice. Moreover, immunogenetic studies among Africans will further our understanding of natural selection and responses to emerging infectious diseases, such as COVID-19.
The scientific imperative of genomic research of African populations is expected to enhance genetic medicine knowledge and practice in Africa but will face the challenges of overburdened and under-resourced public health-care systems, and often absent ethical, legal and social implication frameworks 43 , requiring international collaboration to be managed. Developing an African genomics workforce will be necessary to meet the major need for research across the lifespan for cohorts of millions of individuals with complex or monogenic diseases. Such endeavours can thrive on the foundation of recently established initiatives such as H3Africa. Indeed, equitable access for Africans is essential if African genomics is to reach its full potential as the next frontier of global genetic medicine.
Decoding multifactorial phenotypes
Aravinda Chakravarti. We live in a time of great technological progress in genomics and computing. And we live in a time when ‘genetics’ is a household word, with a public increasingly adept at understanding its relevance to their own lives. Not surprisingly, the study of genetics is being reinvented, rediscovered and reshaped, and we are beginning to understand the science of human heredity at a resolution that was impossible before.
The most significant genetics puzzle today, in my view, is the dissection of ‘family resemblance’ of complex phenotypes, both for intellectual (raison d'être of genetics) and practical (disease diagnosis and therapy) reasons. We have long known that family resemblance arises from shared alleles, declining as genetic relationship wanes, but the precise molecular components and composition of this resemblance are still poorly understood. At the turn of the twentieth century, the components were a matter of bitter and acrimonious debate 44 between the ‘Mendelians’ and the ‘Biometricians’, until the opposing views were reconciled by Ronald Fisher’s 1918 analysis 45 that complex inheritance could be explained through segregation of many genes, each individually Mendelian. In 1920, its publication delayed by World War I, this notion was elegantly demonstrated by the experimental studies of Altenburg and Muller using truncate wing , an “inconstant and modifiable character” 46 in Drosophila .
Fisher’s model assumed an infinite number of genes additively contributing to a trait, with common genetic variation at each component locus comprising two alleles that differ only slightly in their genetic effects 45 ; these genetic assumptions were quite contrary to what was then known 44 . Throughout the past century, this view matured, as segregation analyses of human phenotypes taught us that — beyond the effects of some major genes — most trait variation was polygenic, modulated by family-specific and random environmental factors 47 . Today, we have empirical evidence from GWAS, which use dense maps of genetic variants on hundreds of thousands of individuals measured for many traits and diseases, that the genetic architecture of most multifactorial traits is from common sequence variants with small allelic differences at thousands of sites across the genome 48 . This replacement of a pan-Mendelian view with a pan-polygenic view of traits is one of the most important contributions of genomics to genetics. Unfortunately, this mapping success has not clarified the number of genes involved, the identity of those genes or how those genes specify the phenotype. Indeed, some have concluded that many of the mapped GWAS loci are unrelated to the core biology of each phenotype 49 . Thus, for a deeper understanding, we need radically different approaches to understand complex trait biology in contrast to merely expanding GWAS in larger and larger samples.
for a deeper understanding, we need radically different approaches to understand complex trait biology
Yet, the most significant biology to emerge from GWAS is that most of the likely trait-causing variants fall outside coding sequences, in regulatory elements, most frequently enhancers 50 , 51 . This important finding has uncovered four new genetic puzzles. First, the non-coding regulatory machinery is vast; how much of this regulation is compromised, and how does it affect phenotypes? Second, regulatory changes affect RNA expression at many genes and protein expression at others; how does a cell ‘read’ these numerous changes as specific signals? Third, how is this coordinated expression response translated into cellular responses affecting phenotypes? Fourth, if specific environmental factors affect the same phenotype, which components do they dysregulate? In my opinion, we need to answer these questions for specific traits and diseases to truly understand their polygenic biology. Finally, these explanations must also answer the question of why some traits are decidedly Mendelian whereas others are not.
The questions of tomorrow will need to focus on four areas: the biology of enhancers and the transcription factors that bind them 51 ; the effect of genetic variation in enhancers 50 ; gene regulatory networks (GRNs) that regulate expression of multiple genes 52 ; and how GRN changes lead to specific cellular responses 53 . Despite many advances, the number of enhancers regulating expression of a specific gene remains unknown. How many enhancers are cell type specific versus ubiquitous? How many are constitutive rather than stage specific? And do they act additively or synergistically in gene expression? Additionally, which cognate transcription factors bind these enhancers, with what dynamics and how are they regulated 54 ? These details of a gene’s ‘enhancer code’ are critical for assessing its relative effect on a trait. Next, how does enhancer sequence variation affect a gene’s activity? Does such variation affect transcription factor binding only or its interaction with the promoter? Is the enhancer variant’s effect evident in all cellular states or only some? Is variation in only one enhancer sufficient to alter gene expression, or are multiple changes in multiple elements necessary?
Additional critical questions include which genes are involved in the core pathway underlying a trait, and how do we identify them 49 ? Elegant work has shown how genes are regulated within integrated modular GRNs, whereby one gene’s product is required in a subsequent step by another gene, with feedback interactions 52 . These GRNs comprise elements from the genome, transcriptome and proteome, with rate-limiting steps that require regulation. As our work on Hirschsprung disease has shown 50 , 53 , a GRN is composed of core genes, is the logic diagram of regulation of a major rate-limiting cellular step, is enriched in coding and enhancer disease variants with disease susceptibility scaling with increasing number of variants, and with disease resulting from effects on its rate-limiting gene product 53 . That is, the GRN integrates the expression of multiple genes. Finally, we need to understand how GRN changes alter cell properties and behaviour. I speculate that rate-limiting steps in GRNs are major regulators of broad cell properties, be they differentiation, migration, proliferation or apoptosis, the cellular integrator of GRN variation. Thus, genetic variation across the genome affects enhancers dysregulating many genes, but only when they dysregulate GRNs through rate-limiting steps do they affect cell and tissue biology 55 . This offers the promise of a mechanistic understanding of human polygenic disease.
The way forward for complex trait biology, including disease, is to shift our approach from reverse to forward genetics, using genome-wide approaches to cell type-specific gene perturbation. I believe we can construct cell-type GRNs en masse, inclusive of their enhancers, transcription factors and feedback or feedforward interactions, to then assay functionally defined variation in phenotypes. But, even this approach will be insufficient. We need to test our success by solving at least a few complex traits completely and demonstrating their veracity using a synthetic biology approach to recapitulate the phenotype in a model system; similarly to the field of chemistry, analysis has to be followed by de novo synthesis. Our genomic technologies are getting up to the task to enable this advance; as geneticists, are we?
Enhancers and embryonic development
Eileen Furlong. The work of my group sits at the interface of genome regulation and animal development, and there have been many exciting advances in both during the past decade. Developmental biology studies fundamental processes such as tissue and organ development and how complexity emerges through the combined action of cell communication, movement and mechanical forces. After the discovery that differentiated cells could be reprogrammed to a naive embryonic stem cell-like state, the past decade has witnessed an explosion in in vitro cellular reprogramming and differentiation studies. Organoids are a very exciting extension of this. The extent to which these fairly simple systems can self-organize and generate complexity 56 is one of the unexpected surprises of the past 5–10 years. The buzz around stem cells has also renewed interest in cellular plasticity in vivo and has uncovered an unexpected degree of transdifferentiation and dedifferentiation 57 . In the mouse heart, for example, cardiomyocytes dedifferentiate and proliferate to regenerate heart tissue when damaged within the first week after birth 58 .
Our understanding of the molecular changes that accompany differentiation has hugely advanced owing to the jump in scale, resolution and sensitivity of next-generation sequencing technologies over the past decade. This has led to a flood of studies in embryonic stem cells, iPS cells and embryos that revealed new concepts underlying genome regulation by measuring transcript diversity, transcription factor occupancy, chromatin accessibility and conformation, and chromatin, DNA and RNA modifications. The future challenge will be to connect this information to the physical characteristics of cells and how they form complex tissues. New technologies that solve many challenges of working with embryos will help, including CRISPR to engineer genomes, optogenetics to perturb proteins, lattice light-sheet and selective plane illumination microscopy to image processes in vivo, and low-input methods to overcome issues with scarce material. Particularly exciting to me are recent advances in single-cell genomics, which, although they are in their early days, will dramatically change the way we study embryogenesis. Many new insights have already emerged, including the discovery of unknown cell types and new developmental trajectories for well-established cell types. Even the concept of ‘cell identity’ has come into question.
Cell identities are largely driven by transcription factors, which act through cis -regulatory elements called ‘enhancers.’ One of the most exciting unsolved mysteries, in my opinion, is how enhancers relay information to their target genes. The textbook view of enhancers is of elements with exclusive function that regulate a specific target gene through direct promoter interactions, which occur sequentially if multiple enhancers are involved. However, emerging concepts in the past decade question many of these ‘dogmas’. Some enhancers have dual functions, whereas others may even regulate two genes. Enhancer–promoter communication is now viewed in the light of spatial genome organization, including topologically associating domains (TADs) and membraneless nuclear microcompartments (that is, hubs or condensates) 59 . Being present within the same TAD likely increases the frequency of enhancer–promoter interactions, but how a specific enhancer finds its correct promoter within a TAD, or when TADs are rearranged 60 , 61 , remains a mystery. Hubs or condensates are dynamic microcompartments 62 that contain high local concentrations of proteins, including transcription factors and the transcriptional machinery. One potential implication of condensates is that enhancers may not need to ‘directly’ touch a gene’s promoter to regulate transcription — rather, it may be sufficient to come in close proximity within the same condensate. Presumably, once proteins reach a critical concentration, transcription will be initiated. While this model fits a lot of emerging data, there are still many open questions. What is the required distance between an enhancer and a promoter to trigger transcription? Does this distance differ for different enhancers 63 depending on their transcription factor–DNA affinities? Do different chromatin environments 64 influence the process? At some loci, mutation of a single transcription factor-binding site in a single enhancer can have dramatic effects on gene expression and development. It is difficult to reconcile such cases with a shared condensate model, as other proteins bound to the enhancers and promoter should still phase separate. By contrast, there are many examples where mutation of a single transcription factor-binding site, or even an entire enhancer, has minimal impact on the expression of a gene. These observations suggest that there may be different types of loci, with requirements for different types of chromatin topologies and local nuclear environments, which will be important to tease apart in the coming years.
The genetic dissection of model loci in the 1990s and the first decade of the twenty-first century led to much of our understanding of how genes are regulated. The power of genomics in the past few decades has captured regulatory information for all genes genome-wide, providing more unbiased views of regulatory signatures, leading to new models of gene regulation. What is missing is empirical testing at a large scale. A major challenge is to move to more systematic in vivo functional dissection in organisms. CRISPR-based pooled screens have advanced the interrogation of genomic regions in cell culture systems. However, scaling functional assays in embryos remains a huge challenge. The task is enormous — even long-standing model organisms, such as Drosophila and mice, lack knockout strains for all protein-coding genes, and the number of regulatory elements is at least an order of magnitude higher. There has been little progress in developing scalable methods to quantify the contribution of a transcription factor’s input to an enhancer’s activity, and gene expression, in embryos. More systematic unbiased data will uncover more generalizable regulatory principles, increase our predictive abilities of gene regulation and developmental programmes, and enhance our understanding of the impact of genetic variation.
A major challenge is to move to more systematic in vivo functional dissection in organisms
Perhaps the most promising and exciting prospects in the coming years are to use single-cell genomics, imaging and the integration of the two to dissect the amazing complexity of embryonic development. Single-cell genomics can reveal information about developmental transitions in a way that was unfeasible before. When combined with temporal information, such data can reconstruct developmental trajectories 65 , 66 and identify the regulatory regions and transcription factors likely responsible for each transition 67 . The scale and unbiased nature of the data, profiling tens to hundreds of thousands of cells, provides much richer information than anyone envisaged just 5 years ago, bringing a new level of inference and causal modelling. The ability to measure single-cell parameters in situ (called ‘spatial omics’) will be transformative in the context of developing embryos to reveal the functional impact of spatial gradients, inductive signals and cell–cell interactions, and to move to digital 4D embryos. Combining these approaches with genetic perturbations holds promise to decode developmental programmes as they unfold. Will this bring us to a predictive understanding of the regulatory networks driving embryonic development during the next decade? ‘Simple’ model organisms are a fantastic test case to determine the types and scale of data required and to develop the computational framework to build predictive networks. The systematic functional dissection of gene regulation and true integration of single-cell genomics with single-cell imaging will bring many exciting advances in our understanding of the programmes driving embryonic development in the coming years.
Spatial multi-omics in single cells
Barbara Treutlein. Incredibly, the first single-cell transcriptome was sequenced just over a decade ago 68 ! Since this milestone, transcriptomes of millions of cells have been sequenced and analysed from diverse organisms, tissues and other cellular biosystems, and these maps of cell states are revolutionizing the life sciences. The technologies and associated computational methods have matured and been democratized to such an extent that nearly all laboratories can apply the approach to their particular system or question.
Of course, the transcriptome is not enough, and protocols have already been developed to measure chromatin accessibility, histone modifications, protein abundances, cell lineages and other features linked to genome activity in single cells 69 . Currently, many studies use dissociation-based single-cell genomics methods, where the spatial context is disrupted to facilitate the capture of single cells for downstream processing. Methods are improving to measure genomic features in situ 70 , as well as to computationally map features to spatial contexts 71 , 72 . The stage is set for the next phase of single-cell genomics, where spatial registration of multimodal genome activity across molecular, cellular and tissue or ecosystem scales will enable virtual reconstructions with extraordinary resolution and predictive capacity. These virtual maps will rely on multi-omic profiling of healthy and perturbed tissues and organisms, which presents major challenges and opportunities for innovation.
Cell throughput remains a challenge, and it is unclear what role dissociation-based single-cell sequencing protocols will play in the future. These protocols are fairly easy to implement, and laboratories around the world can execute projects with tens of thousands of cells analysed per experiment. However, there are scenarios in which measuring millions of cells per experiment would be desired, such as in perturbation screens. Combinatorial barcoding methods push cell-throughput boundaries 73 ; however, it is unclear how to scale full transcriptome sequencing economically to millions of cells using current sequencing technologies. ‘Compressed sensing’ modalities — whereby a limited, selected and/or random number of features are measured per cell, and high-dimensional feature levels are recovered through inference or similarity to a known reference — provide an interesting possibility to increasing cell throughput 74 .
Most single-cell transcriptome protocols are currently limited to priming the polyadenylation track present on all cellular mRNAs; however, this approach leads to biased sampling of highly expressed mRNAs. Clever innovations for random or targeted RNA enrichment could be a way to build up composite representations of cell states. Image-based in situ sequencing methods provide a means for increasing the number ofcells measured per experiment, as millions of cells can be imaged without a substantial increase in financial cost, although imaging time is a limiting factor. There remains a lot of room for experimental and computational optimizations to measure the transcriptome, random barcodes, DNA conformations and protein abundances from the micrometre scale to the centimetre scale spatially, and it will be interesting to see how methods for spatial registration advance over the next 5 years.
Currently, most high-throughput measurements are performed on cell suspensions or on intact tissues using one modality. That said, studies are emerging that measure several features from the same cell; for example, mRNA and chromatin accessibility 75 or mRNA and lineage 76 . To build virtual maps, independent measurements from different cells can be integrated with use of data integration tools 77 , although it can be difficult to align cell states across modalities in particular in developing systems. Therefore, the ultimate goal is to directly measure as many features as possible (for example, RNA, lineage, chromatin, proteins and DNA methylation) in the same cell 78 , ideally with spatial resolution. Furthermore, combining genetic and pharmacological perturbation screens with single-cell multi-omic measures will be informative to understand cell state landscapes and underlying regulatory networks for each cell type. The CRISPR–Cas field continues to develop creative tools for precise single-locus editing and other manipulations 79 , and incorporation of these toolkits with single-cell sequencing readouts will certainly bring new mechanistic insight.
Life forms are inherently dynamic, and each cell has a story to tell. Static measurements do not provide sufficient insight into the mechanisms that give rise to each cell state observed in a tissue. Computational approaches to stitch together independent measurements across time can be used to reconstruct potential histories; however, these are indirect inferences. Long-term live imaging in 2D cultures using confocal microscopy and in 3D tissues using light-sheet microscopy provides morphology, behaviour, location and, in some cases, molecular information on the history of a cell. Indeed, such long-term imaging experiments revealed that cell fates or states can be predicted from cell behaviour across many generations 80 . Cell tracking combined with end point single-cell genomics experiments can help to understand how cell states came to be; however, these experiments lack molecular resolution of the intermediates. There are strategies using CRISPR–Cas systems to capture highly prevalent RNAs inside cells at given times and insert these RNAs into DNA for storage and subsequent readout 81 . Together with live tracking and end-point single-cell genomics, such methods could provide unprecedented insight into cell histories.
My vision is that the emerging technologies described above can be applied to human 2D cell culture and 3D organoid biosystems to understand human development and disease mechanisms. My team and others are working to build virtual human organs that are based on high-throughput, multimodal single-cell genomics data. Organoid counterparts provide opportunities to perturb the system and understand lineage histories. Together, the next generation of single-cell genomics methods and human organoid technologies will provide unprecedented opportunities to develop new therapies for human disease.
the next generation of single-cell genomics methods and human organoid technologies will provide unprecedented opportunities
Unravelling the layers of the epigenome
Alexander Meissner. Around 1975, the idea that 5-methylcytosine could provide a mechanism to control gene expression gained traction, despite little knowledge of its genomic distribution or the associated enzymes 82 . With similarly limited genomic information or knowledge of the players involved, the histone code hypothesis was put forward in 2000 to explain how multiple different covalent modifications of chromatin may be coordinated to direct specific regulatory functions 83 . Tremendous progress has been made since, and the list of core epigenetic regulators that have been discovered and characterized seems largely complete 84 .
DNA sequencing has continued to dominate the past decade and contributed to an exponential growth of genome-wide maps of all layers of regulation. In the early days, individual CpG sites could be measured by restriction enzymes, whereas now we have generated probably well over a trillion cytosine methylation measurements. An equally astonishing number of genome-wide data sets have been collected for transcriptomes, histone modifications, transcription factor occupancy and DNA accessibility. Furthermore, the number of single-cell transcriptome and epigenome data sets continues to grow at an unprecedented pace.
On the basis of this overabundance of data across many normal and diseased cell states, for instance, we now clearly understand the non-random distribution of cytosine methylation across many different organisms. These maps have helped to refine our understanding of its relationship to gene expression, including the realization that only a few promoters are normally controlled via this modification, whereas gene bodies are actively targeted, and most dynamic changes occur at distal regulatory sites. Similar insights exist for many core histone modifications, and, in general, we have an improved appreciation of the epigenetic writers, readers and erasers involved. Over the past decade, we have seen substantially integrated and multilayered epigenomic analyses that provide a fairly comprehensive picture of epigenomic landscapes, including their dynamics across development and disease.
Additional innovation is now needed around data access and sharing. As noted, there is certainly no shortage of data, but to enable individual researchers to generate and verify hypotheses quickly improved tools are required to access and browse these data. Over the past decade, large coordinated projects such as ENCODE , the Roadmap Epigenomics Project and Blueprint Epigenome have initiated such efforts, but it remains a reality that data are not at everyone’s fingertips quite yet.
Moreover, despite decades of steady and recently accelerated progress, many important questions remain regarding the molecular coordination and developmental functions of these epigenetic modifications. For instance, cytosine methylation at gene bodies has been preserved for more than a billion years of evolution and yet its precise function is still under investigation. How and why did genomic methylation switch to a global mechanism in vertebrates compared with the selected methylation observed in invertebrates? What is the precise function of this modification in each of its regulatory contexts, and how are its ubiquitously acting enzymes recruited to specific sites in the genome? The latter is particularly timely given recent observations that enhancers, but also some repetitive elements, show ongoing recruitment of both de novo methylation and demethylation activity. Moreover, extraembryonic tissues show redirected activity that shares notable similarities with the long observed altered DNA methylation landscape found across most cancer types 85 . Lastly, it is abundantly clear that DNA methylation is essential for mammalian development; but despite us knowing this for nearly three decades, it is not clear how and why developing knockout embryos die. The specific developmental requirements are also largely true for many histone-modifying enzymes; however, it remains incompletely understood how exactly these modifications interact to support gene regulation.
A decade ago it seemed likely that we would answer questions such as these using newly gained sequencing power as a potent tool for generating hypotheses. However, for the most part, epigenomic analyses have expanded a highly valuable, but still largely descriptive, understanding of numerous epigenetic layers. So one may ask, what is different now and why should we expect to answer these questions in the coming years?
Technological innovation has always played a key role in biology, and some broadly applicable, recent breakthroughs will enable us to drive progress in the coming years. These include the transfer of the bacterial innate immunity CRISPR–Cas system as a universal genome-targeting tool 86 as well as for base editing, epigenome editing and various genome manipulations. Similarly, new fast-acting endogenous protein degradation systems have been developed that further enhance our ability to probe for precise function 87 . The past decade also saw major improvements in imaging technologies as well as cell and molecular biology, moving from the 2D space into the 3D space with both organoid cell culture models 88 and chromosome conformation capture approaches for exploring nuclear organization 89 .
Another major shift included the reappreciation that membraneless organelles are a widespread mechanism of cellular organization 90 . In particular, there have been many advances in our understanding of how condensates form and function, including for transcriptional regulation. Together with known properties of modified histones on DNA and the fact that many epigenetic regulators also contain intrinsically disordered regions, it is reasonable to assume that these physical properties will have a major impact on our understanding of chromatin. Importantly, changes in topology have been linked to disease 91 , and similar connections have been reported recently for condensates 92 . This will likely be an exciting area to follow in the coming years.
there have been many advances in our understanding of how condensates form and function, including for transcriptional regulation
Lastly, our research continues to be more and more reliant on multidisciplinary skills, with mathematics, physics, chemistry and computer science playing an ever-more central role in biology, which will require some rethinking in training and institutional organization to accomplish our goals. Going forward, we will need more functional integration, which in part due to the aforementioned selected discoveries is now very tractable. In particular, more refined perturbation of gene activity, which for many chromatin regulators should be separated into catalytic and regulatory functions, together with readouts at multiple levels of resolution will bring us closer to the insights needed. We recently exemplified this with a pipeline that explores epigenetic regulator mutant phenotypes at single-cell resolution 93 . From these studies, we may be able to understand how epigenetic regulators interact with the environment to influence or protect the organismal phenotype, connecting detailed molecular genetics to classical theories of epigenetic phenomena.
As we approach the 100-year anniversary of the detection of 5-methylcytosine in DNA 94 , it seems we can hope to declare at least for some layers of the epigenome that we fully understand the rules under which they operate. This may enable the exploration of more precise therapeutic interventions, for instance by redirecting chromatin modifiers rather than blocking their universal catalytic activities, which are shared between normal and diseased states. Of course, looking back at predictions made just 10 years ago 95 , one should expect many additional unforeseen advances that are just as difficult to predict now as they were back then.
Long non-coding RNAs: a time to build
Howard Chang. Long non-coding RNAs (lncRNAs) are the dominant transcriptional output of many eukaryotic genomes. Although studies over the past decade have revealed diverse mechanisms and disease implications for many lncRNAs, the vast majority of lncRNAs remain mysterious. The fundamental challenge is that we lack the knowledge to systematically transform lncRNA sequence into function. Progress in the next decade may come from a paradigm shift from ‘reading’ to ‘writing’ lncRNAs.
Gene regulation was once thought to be the exclusive province of proteins. Intense efforts for disease diagnosis and treatment focused almost entirely on protein-coding genes and their products, ignoring the vast majority of the genome. Even at the time of the completion of the Human Genome Project, only a handful of functional lncRNAs were known that silenced the expression of neighbouring genes. Thus, it was widely believed that the genome contained mostly ‘junk’, which sometimes made RNA as transcriptional noise.
The human genome is currently estimated to encode nearly 60,000 lncRNAs, ranging from several hundred to tens of thousands of bases, that apparently do not function by encoding proteins 96 . Studies over the past decade discovered that many lncRNAs act at the interface between chromatin modification machinery and the genome. Specific lncRNAs can act as guides, scaffolds or decoys to control the recruitment of specific chromatin modification enzymes or transcription factors to DNA or their dismissal from DNA 97 . lncRNAs can activate as well as silence genes, and these RNAs can target neighbouring genes as a function of local chromosomal folding (in cis ) or at a distance throughout the genome (in trans ). Detailed dissections of individual lncRNAs have revealed that lncRNAs are composed of modular RNA motifs that enable one lncRNA to connect proteins that read, write or erase specific chromatin marks. These findings have galvanized substantial excitement about lncRNAs; laboratories around the world are now investigating the roles of lncRNAs in diverse systems, ranging from control of flowering time in plants to mutations in human genetic disorders.
Nonetheless, the notable progress to date can be viewed as anecdotal — each lncRNA is its own story. When a new lncRNA sequence is recognized in a genome database or RNA profiling experiment, we are still in the dark about what may happen to the cell or organism (if anything) when the lncRNA is removed. Indeed, efforts to ‘read’ lncRNAs have been the dominant experimental strategy over the past two decades. Systematic efforts in the ENCODE, FANTOM and emerging cell atlas consortia have mapped the transcriptional landscape, transcript isoforms and, more recently, single-cell expression profiles of lncRNAs. These powerful data are now combined with genome-scale CRISPR-based methods to inactivate tens of thousands of lncRNAs, one at a time, to observe possible cell defects 98 , 99 . However, many challenges remain. Positive hits require further exploratory studies to define possible mechanisms of action, and we lack a principled strategy to combine lncRNA knockouts to address genetic redundancy and compensation.
A potentially fruitful and complementary direction is the pivot from ‘reading’ to ‘writing’ long RNA scripts. On the basis of the systematic dissection of RNA sequences and secondary structures in lncRNAs, we and others believe that the information in lncRNAs resembles that on a billboard (in which keywords and catchphrases are repeated) rather than a finely honed legal document (where every comma counts). Small units of RNA shapes are repeated within lncRNAs to build up the meaning in the lncRNA billboard, but these RNA shapes can be rearranged in different orders or locations without affecting meaning. These insights have allowed scientists to recognize lncRNA genes from different species that perform the same function even though the primary sequences bear little similarity 100 . Moreover, investigators were able to strip down lncRNAs to their essential ‘words’, composed of these key repeating shapes and one-tenth the size of the original lncRNA, which still functioned in vivo to control chromatin state over a whole chromosome 100 , 101 . Finally, it is now possible to successfully create synthetic lncRNAs. By adding RNA shapes to carefully chosen RNA templates, investigators are starting to create designer lncRNAs that can regulate chromatin in vivo 100 , suffice to partly rescue the physiological lncRNA gene knockout 102 , or target RNAs to specific cytotopic locations within the cell 103 , 104 .
The shift from reading to writing lncRNAs will challenge us on the technical front, leading to potential transformative technologies. Current technologies for massively parallel reporter gene assays are built on short sequence inserts. A plan to build tens of thousands of synthetic lncRNAs will require accurate long DNA or RNA synthesis. These designer sequences will need to be placed into the appropriate locations in the genome and controlled to have proper developmental expression, splicing pattern and RNA chemical modifications. Landmark studies using the XIST lncRNA, which normally silences the second X chromosome in female cells, to silence the ectopic chromosome 21 in Down syndrome cells highlight the biomedical promise of such an approach 105 .
As the field develops technologies for large-scale creation and testing of synthetic lncRNAs, we can rigorously test our understanding of the information content in the language of RNA sequences and shapes. The next decade promises to be an exciting time for building non-coding RNAs and to create entirely new tools to manipulate gene function for biology and medicine.
FAIR genomics to track tumorigenesis
Núria López-Bigas. Cancer research is one of the fields that has probably benefited the most from the technological and methodological advances of genomics. In the span of less than two decades, the field has witnessed an incredible boost in the generation of cancer genomic, epigenomic and transcriptomic data of patients’ tumours, both in bulk and more recently at the single-cell level. My dream as a cancer researcher is to have a full understanding of the path that cells follow towards tumorigenesis. Which events in the life of an individual, a tissue and a particular cell lead to the malignant transformation of some cells? Of course I do not expect to have a deterministic answer, as this is not a deterministic process. Instead we should aim for a quantitative or probabilistic understanding of the key events that drive tumorigenesis. We have solid epidemiological evidence showing that smoking increases the probability of lung cancer, exposure to the Sun raises the probability of developing melanoma and some anticancer treatments increase the probability of secondary neoplasms. But which specific mechanisms at the molecular and cellular levels influence these increases?
One first clear goal of cancer genomics is to catalogue all genes involved in tumorigenesis across different tissues. Although this is a daunting task, it is actually feasible 106 . By analysing the mutational patterns of genes across tumours, one can identify those with significant deviations from what is expected under neutrality, which indicates that these mutations provide a selective advantage in tumorigenesis and are thus driver mutations. We can imagine a future in which through the systematic analysis of millions of sequenced tumour genomes this catalogue or compendium moves closer and closer to completion. For this to happen, not only do we need genome sequencing to expand — this process is already in motion in research, clinical settings and the pharmaceutical industry — but more importantly the resulting data must be made FAIR (findable, accessible, interoperable and reusable) 107 . To this end, consortia and initiatives that promote, catalyse and facilitate the sharing of genomic data, such as the Beyond 1 Million Genomes consortium, the GA4GH or the cBioPortal for Cancer Genomics , are necessary.
Of note, cataloguing genes and mutations involved in cancer development, albeit a very important first step, is still far from the final goal of understanding how and under which conditions they drive tumorigenesis. Framing cancer development as a Darwinian evolutionary process helps me to navigate the path towards this final objective. As is true of any Darwinian process, its two key features are variation and selection. Thanks to the past 15 years of cancer genomics, we now have a much better grasp of the origin of somatic genetic variation between cells across different tissues. The study of the variability in the number, type and genomic distribution of mutations across tumours provides a window into the life history of cells across the somatic tissues of an individual 108 , 109 . In addition, recent studies sequencing the genome of healthy cells in different tissues 110 , 111 , 112 have shown that mutations accumulate in hundreds and thousands in our cells in normal conditions over time. These studies have also detected positive selection in some genes across healthy tissues. Hence, positive selection is a pervasive process that operates not only in tumorigenesis but also in healthy tissues, where it is a hallmark of somatic development of skin, oesophagus, blood and other tissues. Take, for example, clonal haematopoiesis: it results from a continuous Darwinian evolutionary process in which over time (with age) some haematopoietic cells harbouring mutations in certain blood development genes, such as DNMT3A and TET2 , outcompete other cells in the compartment 113 , 114 . This process is part of normal haematopoietic development. Problems arise only when this process gets out of control, leading to leukaemia in the case of blood, or a malignant tumour in solid tissues. Why is it only in rare cases that this ubiquitous interplay between variation and selection becomes uncontrollable and results in full-blown tumorigenesis? Which events, beside known tumorigenic mutations, drive this process?
we now have a much better grasp of the origin of somatic genetic variation between cells across different tissues
If we have learnt something in recent years, it is that virtually all tumours harbour driver mutations 115 , 116 , 117 , implying that driver genomic events are necessary. However, they are clearly not sufficient for tumorigenesis to occur. So, what are these other triggers of the tumorigenic process? What happens in the lung cells of a smoker or in the haematopoietic cells of a patient treated with chemotherapy that increases their chances to become malignant? Epigenetic modifications and changes in selective constraints, such as evolutionary bottlenecks, for example, at the time of chemotherapy, may be part of the answer.
For the near future, my dream is to see a further increase in FAIR cancer genomics data to help us disentangle the step-by-step game of variation and selection in our tissues that leads to tumorigenesis and likely other ageing-related diseases.

Integrating genomics into medicine
Eran Segal. The past 20 years in genomics have been extraordinary. We developed high-throughput sequencing and learned how to use it to efficiently sequence full genomes and measure gene expression and epigenetic marks at the genome-wide scale and even at the single-cell level 118 . Using these capabilities, we created unprecedented catalogues of novel genomes, functional DNA elements and non-coding RNAs from all kingdoms of life 119 . But — perhaps with the exception of cancer 120 and gene therapy for some monogenic diseases 121 — genomics has yet to deliver on its promise to have an impact on our everyday life. For example, drugs and diagnostics are still being developed in the traditional way, with screening assays to find lead compounds for targets typically arising from animal studies, without involving genomics in any of the steps. Moreover, when the global COVID-19 pandemic hit, the genome of the spreading severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was rapidly sequenced, but why some infected individuals exhibit severe disease and others do not remains unknown.
Indeed, our next challenge is to translate the incredible resources and technologies developed in genomics into an improved understanding of health and disease. This improved understanding should transform the field of medicine to use genomics in its transition to personalized medicine, which promises individualized treatment by targeting the right medication to the right person at the right time on the basis of that person’s unique profile. By continuing to focus on more and more measurements and the creation of more atlases and catalogues, we run the danger of drowning in ever-growing amounts of data and correlative findings. Walking down this path can lead to an endless endeavour, as bulk measurements can always be replaced with single-cell ones, or measures at higher temporal and spatial resolution, across more conditions and wider biological contexts.
Instead, we should use genomics to tackle big unanswered questions such as what causes the variation that we see across people in phenotypes, disease susceptibility and drug responses? What is the relative contribution of genetic, epigenetic, microbiome and environmental factors? How are their effects mediated, and what would be the effect of different interventions? Ultimately, we should strive to use genomics to generate actionable and personalized insights that lead to better health. We are now at an inflexion point in genomics that allows us for the first time to apply it to study human biology and realize these ambitious aims 122 .
At the cellular level, we can use iPS cells from patients to derive cellular models of multiple diseases and prioritize treatments based on measuring both their cellular and molecular response (for example, gene expression and epigenetics) to existing drugs and drug combinations. We can even use massively parallel assays to separately measure the effect of each of tens of thousands of rationally designed mutations, including patient-specific mutations, as we have done, for example, in testing the effect of all clinically identified mutations in TP53 on cellular function 123 . Measuring the molecular effects of directed mutations in genes encoding transcription factors and signalling molecules and in other genes can reveal the underlying pathways and regulatory networks of the disease studied and identify putative therapeutic targets. The application of such approaches to fields that are still poorly understood, such as neurodegenerative diseases, can be particularly impactful.
But we can be much more ambitious and directly profile large cohorts of human individuals using diverse ‘omics’ assays. As molecular changes typically precede clinical disease manifestations, longitudinal measurements coupled with clinical phenotyping have the potential of identifying novel disease diagnostics and therapeutic targets. Indeed, biobanks that track large samples of hundreds of thousands of individuals have recently emerged and are proving highly informative 124 . However, at the molecular level their focus has thus far been on genetics. Technological advances and cost reductions now allow us to obtain much deeper person-specific multi-omic profiles that include transcriptome, proteome, methylome, microbiome, immune system and metabolome measurements. Having these data on the same individual and at multiple time points can reveal which omic layer is more perturbed and informative for each disease and identify associations between molecular markers and disease.
The challenge in using such observational data from human cohorts is to identify which of the associations are causal. One way to address this is to wisely select the nature and type of the associations studied. For example, in working with microbiome data, we can move from analyses at the level of species composition to analyses at the level of SNPs in bacterial genes. Such associations are more specific and more likely to be causal, as in the case of a SNP in the dadH bacterial gene, which correlated with metabolism of the primary medication to treat Parkinson disease and the gut microbiota from patients 125 . Another approach is to use longitudinal measurements and separation of time to emulate target trials from observational data 126 . For example, we can select distinct subsets from the cohort that match on several known risk factors (for example, age or body mass index) but differ on a marker of interest (for example, expression of a gene or presence of an epigenetic mark), and compare future disease onset or progression in these two populations. Similarly, retrospective analysis of baseline multi-omic measurements from participants in randomized clinical trials may identify markers that distinguish responders from non-responders and be used for patient stratification or for identifying additional putative targets.
Ultimately, biomarkers identified from observational cohorts need to be tested in randomized clinical trials to establish causality and assess efficacy. In the case of microbial strains extracted from humans, we may be able to skip animal testing and go directly to human trials. In other cases, such as when human genes are being manipulated, we will need to start with cell culture assays and animal testing before performing clinical trials in humans. However, in all cases, tested omic targets should have already shown associations in human individuals, thus making them more likely to be relevant and succeed in trials, as is the case with drug targets for which genetic evidence links them to the disease 127 .
Beyond these scientific challenges, there is the challenge of engaging the public and diverse ethnic and socio-economic groups to participate in such large-scale multi-omic profiling endeavours even before we can present them with immediate benefits. We can start with incentives in the form of informational summary reports of the data measured and gradually move towards carefully and responsibly conveyed actionable insights as we learn more.
Overcoming the aforementioned challenges is not an easy task, but with the breathtaking advances that genomics has undergone in the past two decades, the time may be right to tackle them. Success can transform genomics from being applied mostly in research settings to having it become an integral and inseparable part of medicine.
CRISPR genome editing enters the clinic
Jin-Soo Kim. In the past several years, genome editing has come of age 128 , in particular because of the repurposing of CRISPR systems. Genomic DNA can be modified in a targeted manner in vivo or in vitro with high efficiency and precision, potentially enabling therapeutic genome editing for the treatment of both genetic and non-genetic diseases. All three types of programmable nucleases developed for genome editing, namely zinc-finger nucleases, transcription activator-like effector nucleases and CRISPR nucleases, are now under clinical investigation. In the next several years, we will be able to learn whether these genome-editing tools will be effective and safe enough to treat patients with an array of diseases, including HIV infection, leukaemia, blood disorders and hereditary blindness, heralding a new era in medicine.
If the history of the development of novel drugs or treatments such as gene therapy and monoclonal antibodies is any guide, the road to therapeutic genome editing is likely to be bumpy but ultimately worth travelling. Key questions related to medical applications of programmable nucleases concern their mode of delivery, specificity, on-target activity and immunogenicity. First, in vivo delivery (or direct delivery into patients) of genes or mRNAs encoding programmable nucleases or preassembled Cas9 ribonucleoproteins can be a challenge, given the large size of these nucleases. Ex vivo (or indirect) delivery is, in general, more efficient than in vivo delivery but is limited to cells from blood or bone marrow, which can be collected with ease, edited in vitro and transfused back into patients. Ongoing developments of nanoparticles and viral vectors are expected to enhance and expand in vivo genome editing in tissues or organs not readily accessible with current delivery systems, such as the brain.
Second, programmable nucleases, including CRISPR nucleases, can cause unwanted on-target and off-target mutations, which may contribute to oncogenesis. Several cell-based and cell-free methods have been developed to identify genome-wide CRISPR off-target sites in an unbiased manner 129 , 130 , 131 . But it remains a challenge to validate off-target activity at sites with low mutation frequencies (less than 0.1%) in a population of cells, owing to the intrinsic error rates of current sequencing technologies. Even at on-target sites, CRISPR–Cas9 can induce unexpected outcomes such as large deletions of chromosomal segments 132 . It will be important to understand the mechanisms behind the unusual on-target activity and to measure and reduce the frequencies of such events.
Last but not least, Cas9 and other programmable nucleases can be immunogenic, potentially causing undesired innate and adaptive immune responses. In this regard, it makes sense that initial clinical trials have focused on ex vivo delivery of Cas9 ribonucleoproteins into T cells or in vivo gene editing in the eye, an immunologically privileged organ. Cas9 epitope engineering or novel Cas9 orthologues derived from non-pathogenic bacteria may avoid some of the immune responses, offering therapeutic modalities for in vivo genome editing in tissues or organs with little or no immune privilege.
Base editing 133 , 134 and prime editing 135 are promising new approaches that may overcome some of the limitations of nuclease-mediated genome editing. Base editors and prime editors are composed of a Cas9 nickase, rather than the wild-type Cas9 nuclease, and a nucleobase deaminase and a reverse transcriptase, respectively. Because a nickase, unlike a nuclease, produces DNA single-strand breaks or nicks, but not double-strand breaks (DSBs), base editors and prime editors are unlikely to induce large deletions at on-target sites and chromosomal rearrangements resulting from non-homologous end joining (NHEJ) repair of concurrent on-target and off-target DSBs. Furthermore, when it comes to gene correction rather than gene disruption, these new types of gene editors are much more efficient and ‘cleaner’ than DSB-producing nucleases because they neither require donor template DNA nor rely on error-prone NHEJ; in human cells, DSBs are preferentially repaired by NHEJ, leading to small insertions or deletions (indels), rather than by homologous recombination involving donor DNA.
Base editors and prime editors are also well suited for germline editing and in utero editing (that is, gene editing in the fetus), which should be done with caution, in full consideration of ethical, legal and societal issues. In principle, CRISPR–Cas9 can be used for the correction of pathogenic mutations in human embryos; however, donor DNA is seldom used as a repair template in human embryos 136 . Recurrent or non-recurrent de novo mutations are responsible for the vast majority of genetic diseases. Cell-free fetal DNA in the maternal blood can be used to detect these de novo mutations in fetuses, which are absent in the parents. Some de novo mutations are manifested even before birth, leading to miscarriage, disability or early death after birth; it is often too late and inefficient to attempt gene editing in newborns. These mutations could be corrected in utero using base editors or prime editors without inducing unwanted indels and without relying on inefficient homologous recombination. Compared with germline editing or preimplantation genetic diagnosis, in utero editing, if proven safe and effective in the future, should be ethically more acceptable because it does not involve the creation or destruction of human embryos.
As promising and powerful as they are, current versions of base editors and prime editors can be further optimized and improved. For instance, Cas9 evolved in microorganisms as a nuclease rather than a nickase. Current Cas9 nickases used for base editing (D10A SpCas9 variant) and prime editing (H840A variant) can be engineered to increase their activities and specificities. In parallel, deaminase and reverse transcriptase moieties in base editors and prime editors, respectively, can be engineered or replaced with appropriate orthologues to increase the efficiency and scope of genome editing. It has been shown that base editors can cause both guide RNA-dependent and guide RNA-independent DNA or RNA off-target mutations, raising concerns for their applications in medicine. Prime editors may also cause unwanted on-target and off-target mutations, which must be carefully studied before moving on to therapeutic applications.
Biomedical researchers are now equipped with powerful tools for genome editing. I expect that these tools will be developed further and applied more broadly in both research and medicine in the coming years.
Collins F. The director of the NIH lays out his vision of the future of medical science. Time https://time.com/5709207/medical-science-age-of-discovery (2019).
The National Academies of Sciences, Engineering, and Medicine Organizing Committee for the International Summit on Human Gene Editing. On human gene editing: international summit statement. The National Academies of Sciences, Engineering, and Medicine https://www.nationalacademies.org/news/2015/12/on-human-gene-editing-international-summit-statement (2015).
Centers for Disease Control and Prevention. COVID-19 in racial and ethnic minority groups. CDC https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/racial-ethnic-minorities.html (2020).
Edwards, F., Lee, H. & Esposito, M. Risk of being killed by police use of force in the United States by age, race–ethnicity, and sex. Proc. Natl Acad. Sci. USA 116 , 16793–16798 (2019).
CAS PubMed PubMed Central Google Scholar
Popejoy, A. B. & Fullerton, S. M. Genomics is failing on diversity. Nature 538 , 161–164 (2016).
Popejoy, A. B. et al. The clinical imperative for inclusivity: race, ethnicity, and ancestry (REA) in genomics. Hum. Mutat. 39 , 1713–1720 (2018).
PubMed PubMed Central Google Scholar
Artiga, S. & Orgera, K. Key facts on health and health care by race and ethnicity. Kaiser Family Foundation https://www.kff.org/report-section/key-facts-on-health-and-health-care-by-race-and-ethnicity-coverage-access-to-and-use-of-care/ (2019).
Armstrong, K., Micco, E., Carney, A., Stopfer, J. & Putt, M. Racial differences in the use of BRCA1/2 testing among women with a family history of breast or ovarian cancer. JAMA 293 , 1729–1736 (2005).
CAS PubMed Google Scholar
Bonham, V. L., Callier, S. L. & Royal, C. D. Will precision medicine move us beyond race? N. Engl. J. Med. 374 , 2003–2005 (2016).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581 , 434–443 (2020).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50 , 1219–1224 (2018).
The SIGMA Type 2 Diabetes Consortium. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 506 , 97–101 (2014).
Google Scholar
Wetterstrand, K. A. DNA sequencing costs: data from the NHGRI genome sequencing program (GSP). National Human Genome Research Institute https://www.genome.gov/sequencingcostsdata (2019).
Wall, J. D. et al. The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature 576 , 106–111 (2019).
CAS Google Scholar
Kowalski, M. H. et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 15 , e1008500 (2019).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562 , 203–209 (2018).
Gaziano, J. M. et al. Million veteran program: a mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 70 , 214–223 (2016).
PubMed Google Scholar
Nagai, A. et al. Overview of the BioBank Japan Project: study design and profile. J. Epidemiol. 27 , S2–S8 (2017).
Denny, J. C. et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 31 , 1102–1110 (2013).
Sirugo, G., Williams, S. M. & Tishkoff, S. A. The missing diversity in human genetic studies. Cell 177 , 1080 (2019).
Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51 , 584–591 (2019).
McQuillan, M. A., Zhang, C., Tishkoff, S. A. & Platt, A. The importance of including ethnically diverse populations in studies of quantitative trait evolution. Curr. Opin. Genet. Dev. 62 , 30–35 (2020).
Sohail, M. et al. Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. eLife 8 , e39702 (2019).
Crawford, N. G. et al. Loci associated with skin pigmentation identified in African populations. Science 358 , eaan8433 (2017).
Gasperini, M., Tome, J. M. & Shendure, J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat. Rev. Genet. 21 , 292–310 (2020).
Racimo, F., Sankararaman, S., Nielsen, R. & Huerta-Sánchez, E. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 16 , 359–371 (2015).
Skoglund, P. & Mathieson, I. Ancient genomics of modern humans: the first decade. Annu. Rev. Genomics Hum. Genet. 19 , 381–404 (2018).
Vicente, M. & Schlebusch, C. M. African population history: an ancient DNA perspective. Curr. Opin. Genet. Dev. 62 , 8–15 (2020).
Sherman, R. M. et al. Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat. Genet. 51 , 30–35 (2019).
Durvasula, A. et al. Recovering signals of ghost archaic introgression in African populations. Sci. Adv. 12 , eaax5097 (2020).
Skov, L. et al. The nature of Neanderthal introgression revealed by 27,566 Icelandic genomes. Nature 582 , 78–83 (2020).
Adeyemo, A. A. et al. ZRANB3 is an African-specific type 2 diabetes locus associated with beta-cell mass and insulin response. Nat. Commun. 10 , 3195 (2019).
Cohen, J. et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9 . Nat. Genet. 37 , 161–165 (2005).
Gurdasani, D. et al. Uganda genome resource enables insights into population history and genomic discovery in Africa. Cell 179 , 984–002.e36 (2019).
Gurdasani, D. et al. Genomics of disease risk in globally diverse populations. Nat. Rev. Genet. 20 , 520–535 (2019).
Gulsuner, S. et al. Genetics of schizophrenia in the South African Xhosa. Science 367 , 569–573 (2020).
Shriner, D. & Rotimi, C. N. Whole-genome-sequence-based haplotypes reveal single origin of the sickle allele during the Holocene wet phase. Am. J. Hum. Genet. 102 , 547–556 (2018).
Wu, Y. et al. Highly efficient therapeutic gene editing of human haematopoietic stem cells. Nat. Med. 25 , 776–783 (2019).
Geard, A. et al. Clinical and genetic predictors of renal dysfunctions in sickle cell anaemia in Cameroon. Br. J. Haematol. 178 , 629–639 (2017).
Lebeko, K. et al. Targeted genomic enrichment and massively parallel sequencing identifies novel nonsyndromic hearing impairment pathogenic variants in Cameroonian families. Clin. Genet. 90 , 288–290 (2016).
Genovese, G. et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329 , 841–845 (2010).
Sierra, B. et al. OSBPL10, RXRA and lipid metabolism confer African-ancestry protection against dengue haemorrhagic fever in admixed Cubans. PLoS Pathog. 13 , e1006220 (2017).
Wonkam, A. & de Vries, J. Returning incidental findings in African genomics research. Nat. Genet. 52 , 17–20 (2020).
Provine, W. B. The Origins of Theoretical Population Genetics (University of Chicago Press, 1971)
Fisher, R. A. The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 52 , 399–433 (1918).
Altenburg, E. & Muller, H. J. The genetic basis of truncate wing – an inconstant and modifiable character in Drosophila. Genetics 5 , 1–59 (1920).
Morton, N. E. Analysis of family resemblance. I. Introduction. Am. J. Hum. Genet. 26 , 318–330 (1974).
Visscher, P. M. et al. 10 Years of GWAS discovery: biology, function and translation. Am. J. Hum. Genet. 101 , 5–22 (2017).
Boyle, E. A., Li, Y. I. & Pritchard, J. K. An expanded view of complex traits: from polygenic to omnigenic. Cell 169 , 1177–1186 (2017).
Emison, E. S. et al. A common, sex-dependent mutation in a putative RET enhancer underlies Hirschsprung disease susceptibility. Nature 434 , 857–863 (2005).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337 , 1190–1195 (2012).
Davidson, E. Emerging properties of animal gene regulatory networks. Nature 468 , 911–920 (2010).
Chatterjee, S. et al. Enhancer variants synergistically drive dysregulation of the RET gene regulatory network in Hirschsprung disease. Cell 167 , 355–368 (2016).
Segal, E., Raveh-Sadka, T., Schroeder, M., Unnerstall, U. & Gaul, U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451 , 535–540 (2008).
Chakravarti, A. & Turner, T. N. Revealing rate-limiting steps in complex disease biology: The crucial importance of studying rare, extreme-phenotype families. Bioessays 38 , 578–586 (2016).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501 , 373–379 (2013).
Rothman, J. & Jarriault, S. Developmental plasticity and cellular reprogramming in caenorhabditis elegans. Genetics 213 , 723–757 (2019).
Porrello, E. R. et al. Transient regenerative potential of the neonatal mouse heart. Science 331 , 1078–1080 (2011).
Mir, M., Bickmore, W., Furlong, E. E. M. & Narlikar, G. Chromatin topology, condensates and gene regulation: shifting paradigms or just a phase? Development 146 , dev182766 (2019).
Ghavi-Helm, Y. et al. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 51 , 1272–1282 (2019).
Despang, A. et al. Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat. Genet. 51 , 1263–1271 (2019).
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. & Sharp, P. A. A phase separation model for transcriptional control. Cell 169 , 13–23 (2017).
Shrinivas, K. et al. Enhancer features that drive formation of transcriptional condensates. Mol. Cell 75 , 549–561 e547 (2019).
Narlikar, G. J. Phase-separation in chromatin organization. J. Biosci. 45 , 5 (2020).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566 , 496–502 (2019).
Farrell, J. A. et al. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360 , eaar3131 (2018).
Cusanovich, D. A. et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555 , 538–542 (2018).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6 , 377–382 (2009).
Camp, J. G., Platt, R. & Treutlein, B. Mapping human cell phenotypes to genotypes with single-cell genomics. Science 365 , 1401–1405 (2019).
Lein, E., Borm, L. E. & Linnarsson, S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358 , 64–69 (2017).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33 , 495–502 (2015).
Achim, K. et al. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat. Biotechnol. 33 , 503–509 (2015).
Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357 , 661–667 (2017).
Cleary, B., Cong, L., Cheung, A., Lander, E. S. & Regev, A. Efficient generation of transcriptomic profiles by random composite measurements. Cell 171 , 1424–1436 e1418 (2017).
Cao, J. et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361 , 1380–1385 (2018).
Kester, L. & van Oudenaarden, A. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 23 , 166–179 (2018).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20 , 257–272 (2019).
Zhu, C., Preissl, S. & Ren, B. Single-cell multimodal omics: the power of many. Nat. Methods 17 , 11–14 (2020).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol . (2020).
Loeffler, D. et al. Asymmetric lysosome inheritance predicts activation of haematopoietic stem cells. Nature 573 , 426–429 (2019).
Schmidt, F., Cherepkova, M. Y. & Platt, R. J. Transcriptional recording by CRISPR spacer acquisition from RNA. Nature 562 , 380–385 (2018).
Holliday, R. & Pugh, J. E. DNA modification mechanisms and gene activity during development. Science 187 , 226–232 (1975).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403 , 41–45 (2000).
Jambhekar, A., Dhall, A. & Shi, Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 20 , 625–641 (2019).
Smith, Z. D. et al. Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature 549 , 543–547 (2017).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 , 816–821 (2012).
Nabet, B. et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14 , 431–441 (2018).
Clevers, H. Modeling development and disease with organoids. Cell 165 , 1586–1597 (2016).
Dekker, J., Marti-Renom, M. A. & Mirny, L. A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14 , 390–403 (2013).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18 , 285–298 (2017).
Lupianez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161 , 1012–1025 (2015).
Basu, S. et al. Unblending of transcriptional condensates in human repeat expansion disease. Cell 181 , 1062–1079 e1030 (2020).
Grosswendt, S. et al. Epigenetic regulator function through mouse gastrulation. Nature 584 , 102–108 (2020).
Johnson, T. B. & Coghill, R. D. Researches on pyrimidines. C111. The discovery of 5-methyl-cytosine in tuberculinic acid, the nucleic acid of the tubercle bacillus. J. Am. Chem. Soc. 47 , 2838–2844,47 (1925).
Heard, E. et al. Ten years of genetics and genomics: what have we achieved and where are we heading? Nat. Rev. Genet. 11 , 723–733 (2010).
Quinn, J. J. & Chang, H. Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17 , 47–62 (2016).
Kopp, F. & Mendell, J. T. Functional classification and experimental dissection of long noncoding RNAs. Cell 172 , 393–407 (2018).
Liu, S. J. et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355 , eaah7111 (2017).
Rubin, A. J. et al. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. Cell 176 , 361–376.e17 (2019).
Quinn, J. J. et al. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes. Dev. 30 , 191–207 (2016).
Kirk, J. M. et al. Functional classification of long non-coding RNAs by k-mer content. Nat. Genet. 50 , 1474–1482 (2018).
Carter, A. C. et al. Spen links RNA-mediated endogenous retrovirus silencing and X chromosome inactivation. eLife 9 , e54508 (2020).
Lubelsky, Y. & Ulitsky, I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 555 , 107–111 (2018).
Shukla, C. J. et al. High-throughput identification of RNA nuclear enrichment sequences. EMBO J. 37 , e98452 (2018).
Czerminski, J. T. & Lawrence, J. B. Silencing Trisomy 21 with XIST in neural stem cells promotes neuronal differentiation. Dev. Cell 52 , 294–308 e3 (2020).
Martínez-Jiménez, F. et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer https://doi.org/10.1038/s41568-020-0290-x (2020).
Article PubMed Google Scholar
Wilkinson, M. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3 , 160018 (2016).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578 , 94–101 (2020).
Gonzalez-Perez, A., Radhakrishnan, S. & Lopez-Bigas, N. Local determinants of the mutational landscape of the human genome. Cell 177 , 101–114 (2019).
Martincorena, I. et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348 , 880–886 (2015).
Martincorena, I. et al. Somatic mutant clones colonize the human esophagus with age. Science 362 , 911–917 (2018).
Yokoyama, A. et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 565 , 312–317 (2019).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371 , 2477–2487 (2014).
Jaiswal, S. et al. Age related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371 , 2488–2498 (2014).
Sabarinathan, R. et al. The whole-genome panorama of cancer drivers. Preprint at bioRxiv https://doi.org/10.1101/190330 (2017).
Pich, O. et al. The mutational footprints of cancer therapies. Nat. Genet. 51 , 1732–1740 (2019).
Campbell, P. J. et al. Pan-cancer analysis of whole genomes. Nature 578 , 82–93 (2020).
Rozenblatt-Rosen, O., Stubbington, M. J. T., Regev, A. & Teichmann, S. A. The Human Cell Atlas: from vision to reality. Nature 550 , 451–453 (2017).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489 , 57–74 (2012).
Damodaran, S. et al. Cancer Driver Log (CanDL): catalog of potentially actionable cancer mutations. J. Mol. Diagn. 17 , 554–559 (2015).
High, K. A. & Roncarolo, M. G. Gene therapy. N. Engl. J. Med. 381 , 455–464 (2019).
Shilo, S., Rossman, H. & Segal, E. Axes of a revolution: challenges and promises of big data in healthcare. Nat. Med. 26 , 29–38 (2020).
Kotler, E. et al. A systematic p53 mutation library links differential functional impact to cancer mutation pattern and evolutionary conservation. Mol. Cell 71 , 873 (2018).
Swanson, J. M. The UK Biobank and selection bias. Lancet 380 , 110 (2012).
Maini Rekdal, V., Bess, E. N., Bisanz, J. E., Turnbaugh, P. J. & Balskus, E. P. Discovery and inhibition of an interspecies gut bacterial pathway for levodopa metabolism. Science 364 , eaau6323 (2019).
Hernán, M. A. & Robins, J. M. Using big data to emulate a target trial when a randomized trial is not available. Am. J. Epidemiol. 183 , 758–764 (2016).
Nelson, M. R. et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 47 , 856–860 (2015).
Kim, J.-S. Genome editing comes of age. Nat. Protoc. 11 , 1573–1578 (2016).
Kim, D. et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12 , 237–243 (2015).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33 , 187–197 (2015).
Wienert, B. et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364 , 286–289 (2019).
Kosicki, M., Tomberg, K. & Bradley, A. et al. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36 , 765–771 (2018).
Komor, A. C. et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533 , 420–424 (2016).
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353 , aaf8729 (2016).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 , 149–157 (2019).
Ma, H. et al. Correction of a pathogenic gene mutation in human embryos. Nature 548 , 413–419 (2017).
Download references
Acknowledgements
A.C. acknowledges that the ideas in his contribution were developed through studies on Hirschsprung disease and thanks the many trainees who have contributed to this work over the past 5 years. A.L.M. acknowledges A. Gutierrez, K. Kostick, G. Lazaro, M. Majumder, K. Munoz, S. Pereira, H. Smith and P. Zuk for feedback. A.M. thanks D. Hnisz, Z. D. Smith, J. Charlton and H. Kretzmer for feedback and the Max Planck Society for funding. A.W. is supported by NIH awards U54HG009790, U01HG009716, U01HG007459 and U24HL135600, and Wellcome Trust award H3A/18/001, and states that the funders had no role in study design, and analysis, decision to publish or preparation of the manuscript. B.T. acknowledges J. G. Camp for helpful discussions. E.E.M.F. is very grateful to A. Ephrussi, M. Mir, M. Perino, Y. Kherdjemil, T. Pollex and S. Secchia for useful comments. E. E. M. F is supported by European Research Council (Advanced Grant) agreement no. 787611 (DeCRyPT). E.S. is supported by grants from the European Research Council and the Israel Science Foundation. H.Y.C. is supported by NIH RM1-HG007735 and R35-CA209919. H.Y.C. is an investigator of the Howard Hughes Medical Institute. J.-S.K. is supported by the Institute for Basic Science (IBS-R021-D1). N.L-B. acknowledges funding from the European Research Council (Consolidator Grant 682398), the Spanish Ministry of Economy and Competitiveness (SAF2015-66084-R, European Regional Development Fund) and the Asociación Española Contra el Cáncer (GC16173697BIGA). S.A.T. is funded by NIH grants R35 GM134957-01 and NIAMS R01AR076241-01A1 and American Diabetes Association Pathway to Stop Diabetes grant #1-19-VSN-02.
Author information
Authors and affiliations.
Center for Medical Ethics and Health Policy, Baylor College of Medicine, Houston, TX, USA
Amy L. McGuire
Broad Institute of MIT and Harvard, Cambridge, MA, USA
Stacey Gabriel & Alexander Meissner
Department of Genetics, University of Pennsylvania, Philadelphia, PA, USA
Sarah A. Tishkoff
Department of Biology, University of Pennsylvania, Philadelphia, PA, USA
Department of Medicine, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
Ambroise Wonkam
Institute of Infectious Diseases and Molecular Medicine, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
Center for Human Genetics and Genomics, New York University Grossman School of Medicine, New York, NY, USA
Aravinda Chakravarti
European Molecular Biology Laboratory, Genome Biology Department, Heidelberg, Germany
Eileen E. M. Furlong
Department of Biosystems Science and Engineering, ETH Zurich, Basel, Switzerland
Barbara Treutlein
Department of Genome Regulation, Max Planck Institute for Molecular Genetics, Berlin, Germany
Alexander Meissner
Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA, USA
Institute of Chemistry and Biochemistry, Freie Universität Berlin, Berlin, Germany
Center for Personal Dynamic Regulomes, Howard Hughes Medical Institute, Stanford University, Stanford, CA, USA
Howard Y. Chang
Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain
Núria López-Bigas
Research Program on Biomedical Informatics, Universitat Pompeu Fabra, Barcelona, Spain
Institució Catalana de Recerca i Estudis Avançats, Barcelona, Spain
Department of Computer Science and Applied Mathematics, Weizmann Institute of Science, Rehovot, Israel
Center for Genome Engineering, Institute for Basic Science, Daejon, Republic of Korea
Jin-Soo Kim
You can also search for this author in PubMed Google Scholar
Corresponding authors
Correspondence to Amy L. McGuire , Stacey Gabriel , Sarah A. Tishkoff , Ambroise Wonkam , Aravinda Chakravarti , Eileen E. M. Furlong , Barbara Treutlein , Alexander Meissner , Howard Y. Chang , Núria López-Bigas , Eran Segal or Jin-Soo Kim .
Ethics declarations
Competing interests.
H.Y.C. is a co-founder of Accent Therapeutics and Boundless Bio and an advisor of 10x Genomics, Arsenal Biosciences and Spring Discovery. J.-S.K. is a co-founder of and holds stock in ToolGen Inc. A.C., A.L.M., A.M., A.W., B.T., E.E.M.F., E.S., N.L.-B., S.G. and S.A.T. declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Beyond 1 Million Genomes : https://b1mg-project.eu/
Blueprint Epigenome : https://www.blueprint-epigenome.eu/
cBioPortal for Cancer Genomics : https://www.cbioportal.org/
ENCODE : https://www.encodeproject.org/
Global Alliance for Genomics and Health : https://www.ga4gh.org/
gnomAD : https://gnomad.broadinstitute.org/
GTEx : https://www.gtexportal.org/home/
GWAS Catalog : https://www.ebi.ac.uk/gwas
H3Africa : https://h3africa.org
Roadmap Epigenomics Project : http://www.roadmapepigenomics.org/
Rights and permissions
Reprints and permissions
About this article
Cite this article.
McGuire, A.L., Gabriel, S., Tishkoff, S.A. et al. The road ahead in genetics and genomics. Nat Rev Genet 21 , 581–596 (2020). https://doi.org/10.1038/s41576-020-0272-6
Download citation
Accepted : 21 July 2020
Published : 24 August 2020
Issue Date : October 2020
DOI : https://doi.org/10.1038/s41576-020-0272-6
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Identifying knowledge deficiencies in genetics education among medical students and interns in saudi arabia- a cross-sectional study.
- Abeer F. Zakariyah
- Sadin A. Alamri
- Mehenaz A. Hanbazazh
BMC Medical Education (2024)
VUS next in rare diseases? Deciphering genetic determinants of biomolecular condensation
- María Heredia-Torrejón
- Raúl Montañez
- Alfonso M. Lechuga-Sancho
Orphanet Journal of Rare Diseases (2024)
Overcoming barriers to single-cell RNA sequencing adoption in low- and middle-income countries
- Tracy Boakye Serebour
- Adam P. Cribbs
- Sarah J. B. Snelling
European Journal of Human Genetics (2024)
An overview of artificial intelligence in the field of genomics
- Khizra Maqsood
- Hani Hagras
- Nicolae Radu Zabet
Discover Artificial Intelligence (2024)
IMA Genome – F19
- Janneke Aylward
- Andi M. Wilson
- Brenda D. Wingfield
IMA Fungus (2024)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.
- Search Menu
- Sign in through your institution
- Advance Articles
- Perspectives
- Knowledgebase and Database Resources
- Nobel Laureates Collection
China Virtual Outreach Webinar
Neurogenetics.
- Fungal Genetics and Genomics
- Multiparental Populations
- Genomic Prediction
- Plant Genetics and Genomics
Genetic Models of Rare Diseases
- Genomic Data Analyses In Biobanks
Genetics of Bacteria
- Why Publish
- Author Guidelines
- Submission Site
- Open Access Options
- Full Data Policy
- Self-Archiving Policy
- About Genetics
- About Genetics Society of America
- Editorial Board
- Early Career Reviewers
- Guidelines for Reviewers
- Advertising & Corporate Services
- Journals on Oxford Academic
- Books on Oxford Academic

Editor-in-Chief
Howard Lipshitz
Executive Editor
Tracey DePellegrin
Managing Editor
Ruth Isaacson
Scientific Editor and Program Manager
Genetics of bacteria: a call for papers.
The journals of the Genetics Society of America, GENETICS and G3: Genes|Genomes|Genetics, are calling for submissions of papers on the Genetics of Bacteria. The submission deadline is December 2, 2024.
Why publish with GENETICS?
Why publish in genetics.
Learn more about why GENETICS is the perfect home for your research, and submit today to join our celebrated author community.
Why publish?
Series and Collections accepting papers
Submit your work to one of GSA’s ongoing series and collections.
Currently accepting submissions
Meet the Editorial Board
See who handles papers for GENETICS by topic.
Editorial board
Re-watch the recent China Virtual Outreach Webinar where you will learn more about publishing your work in the journal.
Watch the webinar
Latest articles
Series & collections.

Classical genetic analyses of bacteria have made numerous and profound contributions to both fundamental and applied science. From early studies on gene regulation and the nature of horizontal gene transfer, through to the recent discovery and harnessing of CRISPR, bacterial genetics continues to yield important advances. Genetic and molecular studies on DNA replication, transcription, genome stability, and mutagenesis have provided insights into essential cellular and evolutionary processes.
This series will highlight recent genetic and genomic work that leverages both classical and innovative genetic approaches in the study of bacterial genetics.

Genes and variants of interest in rare diseases often benefit from modelling in cellular assays or genetic models to aid in understanding molecular and cellular mechanisms of disfunction. Model organisms are useful for the discovery of new genetic diseases and key to understanding variant effects, and modelling a disease gene in a genetic model means that researchers can perform an in-depth exploration of gene or variant function. The GSA Journals are pleased to publish a series highlighting ongoing advances in rare disease discovery and mechanisms by presenting key research findings and new discoveries.

Plant Genetics and Genomics
Plant science has generated many discoveries and advances in genetics and genomics research. These contributions reflect the ingenuity and rigor of the plant science community, as well as the rich diversity of plants and their biology. To showcase this critical work, GENETICS and G3: Genes|Genomes|Genetics has launched the Plant Genetics and Genomics series with a collection of fourteen research articles and an accompanying editorial.

Neurogenetics lies at the intersection of Neuroscience and Genetics, where genetic approaches are applied to the study of nervous system development, function, and plasticity. Overseen by Series Editors Oliver Hobert, Cecilia Moens, and Kate O’Connor Giles, this new series aims to make the GSA Journals a home for cutting-edge, robust research in neurogenetics.

FlyBook from GENETICS is a comprehensive compendium of review articles presenting the current state of knowledge in Drosophila research.
Browse FlyBook

WormBook from GENETICS features a comprehensive compendium of review articles presenting the current state of knowledge in C. elegans research. WormBook articles will span the breadth of the biology, genetics, genomics, and evolutionary biology of C. elegans .
Browse WormBook

The YeastBook series from GENETICS features a comprehensive compendium of reviews that presents the current state of knowledge of the molecular biology, cellular biology, and genetics of the yeast Saccharomyces cerevisiae .
Browse YeastBook
More from GSA

G3: Genes|Genomes|Genetics
G3, a Genetics Society of America journal, provides a forum for the publication of high-quality foundational research-particularly research that generates useful genetic and genomic information, as well as genome reports, mutant screens, and advances in methods and technology.
Find out more

GSA members of all career stages receive member benefits including access to professional development programs, discounted meeting registration, and eligibility for travel awards. Members also receive a personal subscription to GENETICS, as well as discounted publication fees in both GSA journals.

Conferences
GSA conferences have long served as community hubs for researchers focused on particular organisms or topics. GSA also hosts The Allied Genetics Conference (TAGC) , a unique meeting that brings together multiple research communities for collaboration and synthesis.
Attend a conference

Career Development
GSA professional development programs provide rich opportunities for scientists to gain skills, experience, mentors, and networks. Our initiatives and resources range from peer review training to inclusive public engagement, newsletters, webinars, a job board, leadership programs, and much more.
Browse Opportunities

Email alerts
Register to receive email alerts as soon as new content from GENETICS is published online.

Recommend to your library
Fill out our simple online form to recommend GENETICS to your library. Recommend now

Author resources
Learn about how to submit your article, our publishing process, and tips on how to promote your article.
Related Titles
- Recommend to Your Librarian
- Advertising and Corporate Services
- Journals Career Network
Affiliations
- Online ISSN 1943-2631
- Copyright © 2024 Genetics Society of America
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
The Study of Genetics: Importance for Society Essay
Scientific progress does not stand still but instead constantly stimulates the development of social thought. Thanks to the products of scientific progress, people already use dozens of electronic devices and technological solutions on a daily basis to optimize life. One of the fundamental areas of this development is genetic research. Thus, the study of genetics makes it possible to solve pressing issues related to medicine, forensics, and security. This essay aims to discuss the public importance of supporting genetic research.
With the advances of the genetic sciences, medical progress has reached severe results. A key advantage of genetics for the clinical environment is the real possibility of manipulating the genetic code of a patient’s DNA in order to edit it. Adjustments in the polynucleotide sequences are intended to solve the problem of hereditary diseases and to defeat the harmful, damaging mutations that degrade the quality of life of millions of people. Through genetic manipulation, humanity is expected to be able to solve cancer, type I diabetes, and even schizophrenia (Vincent & Yaghootkar, 2020; Zhuo et al., 2017). Consequently, the benefit of studying genetics for medicine cannot be denied.
On the other hand, encouraging genetic research optimizes forensic systems. By the current moment, many laboratories are using genetics to identify suspects accurately. If traces of the killer are left at the scene of violent crimes, it is possible to qualitatively study their DNA and, consequently, compare it to that of suspects using PCR and electrophoresis techniques (Sebastiana, 2021). In other words, genetic studies simplify the work of law enforcement. So it should be understood that by the same scheme, it is possible to establish paternity, which simplifies the system of legal proceedings and gives reliable results regarding the relationship between the child and the man. Thus, for forensics and genealogy, the use of genetic methods is critical.
Finally, the use of genetics for public safety is of great importance. It should be understood that, in this context, security implies food security because one day, humanity will come to lose sufficient food resources. In such a case, genetic science makes it possible to create genetically modified products, which have a competitive advantage over natural plant forms. By editing the genome of plants, humans can modify their economic properties and thus increase their viability, sustainability, and fertility. That said, the academic community is unequivocally inclined to consider GMOs as utterly safe as conventional food (Biddle, 2018). To put it another way, the use of genetic science has extremely significant implications for the agricultural industry as well. As a result, it allows for the food security of communities in case natural foods are lost.
Finally, it should be emphasized that the promotion of genetic research is directly linked to the improvement of the quality of life of society. Humanity cannot live by conservative rules and abandon scientific and technological progress achievements: such views have a destructive effect. Consequently, the study of genetics is vital for the sustainability of society. As has been shown in this essay, genetics is fundamentally important for medicine, forensics, genealogy, and the agricultural sector. Indeed, the range of functional benefits of genetics is considerably broader. All of this together leads to the conclusion that the genetic sciences must be sustained and that research in these areas is essential to all of society, both in the short and long term.
Biddle, J. B. (2018). “Antiscience Zealotry”? Values, epistemic risk, and the GMO debate. Philosophy of Science, 85 (3), 360-379.
Sebastiana, M. (2021). PCR – polymerase chain reaction. Biologia Vegetal , 1-6.
Vincent, E. E., & Yaghootkar, H. (2020). Using genetics to decipher the link between type 2 diabetes and cancer: Shared aetiology or downstream consequence? Diabetologia, 63 (9), 1-12.
Zhuo, C., Hou, W., Hu, L., Lin, C., Chen, C., & Lin, X. (2017). Genomic editing of non-coding RNA genes with CRISPR/Cas9 ushers in a potential novel approach to study and treat schizophrenia. Frontiers in Molecular Neuroscience, 10 , 28-35.
- Adaptation and Natural Selection
- Bioinformatics of Enzyme Kinase
- Modern Technology in DNA and Genealogy Solving Cold Case Murders
- "A Genealogy of Modern Racism" by C. West
- The Controversy of Darwin’s Theory
- The Development of the Neural System and Genetic Program
- Genetic Disorder: “A Genetic Link to Anorexia”
- Genetic Enhancement: Ethical Aspect
- Genetic Modification and Implicit Bias Against People With Disabilities
- Genetics and Genomics in Healthcare
- Chicago (A-D)
- Chicago (N-B)
IvyPanda. (2022, August 27). The Study of Genetics: Importance for Society. https://ivypanda.com/essays/the-study-of-genetics-importance-for-society/
"The Study of Genetics: Importance for Society." IvyPanda , 27 Aug. 2022, ivypanda.com/essays/the-study-of-genetics-importance-for-society/.
IvyPanda . (2022) 'The Study of Genetics: Importance for Society'. 27 August.
IvyPanda . 2022. "The Study of Genetics: Importance for Society." August 27, 2022. https://ivypanda.com/essays/the-study-of-genetics-importance-for-society/.
1. IvyPanda . "The Study of Genetics: Importance for Society." August 27, 2022. https://ivypanda.com/essays/the-study-of-genetics-importance-for-society/.
Bibliography
IvyPanda . "The Study of Genetics: Importance for Society." August 27, 2022. https://ivypanda.com/essays/the-study-of-genetics-importance-for-society/.
- To find inspiration for your paper and overcome writer’s block
- As a source of information (ensure proper referencing)
- As a template for you assignment
IvyPanda uses cookies and similar technologies to enhance your experience, enabling functionalities such as:
- Basic site functions
- Ensuring secure, safe transactions
- Secure account login
- Remembering account, browser, and regional preferences
- Remembering privacy and security settings
- Analyzing site traffic and usage
- Personalized search, content, and recommendations
- Displaying relevant, targeted ads on and off IvyPanda
Please refer to IvyPanda's Cookies Policy and Privacy Policy for detailed information.
Certain technologies we use are essential for critical functions such as security and site integrity, account authentication, security and privacy preferences, internal site usage and maintenance data, and ensuring the site operates correctly for browsing and transactions.
Cookies and similar technologies are used to enhance your experience by:
- Remembering general and regional preferences
- Personalizing content, search, recommendations, and offers
Some functions, such as personalized recommendations, account preferences, or localization, may not work correctly without these technologies. For more details, please refer to IvyPanda's Cookies Policy .
To enable personalized advertising (such as interest-based ads), we may share your data with our marketing and advertising partners using cookies and other technologies. These partners may have their own information collected about you. Turning off the personalized advertising setting won't stop you from seeing IvyPanda ads, but it may make the ads you see less relevant or more repetitive.
Personalized advertising may be considered a "sale" or "sharing" of the information under California and other state privacy laws, and you may have the right to opt out. Turning off personalized advertising allows you to exercise your right to opt out. Learn more in IvyPanda's Cookies Policy and Privacy Policy .
2022 DNA Day Essay Contest: Full Essays
1 st Place : Man Tak Mindy Shie, Grade 12 Teacher: Dr. Siew Hwey Alice Tan School: Singapore International School (Hong Kong) Location: Hong Kong, China
Many would say that the most significant stride in recent genetics has been the completion of the human genome, which laid the basis for studying genetic variation. However, let us not forget that this began with the understanding of heredity based on Gregor Mendel’s observations in 1857.
Observations from Mendel’s pea plant hybridization experiments led to two fundamental principles of inheritance (1). The first was the Law of Segregation, which states that reproductive gamete cells transmit only one allele to their offspring. This means that a diploid offspring will inherit one allele from each parent. We now understand genes to be the units of heredity that carry genetic information and alleles to be different variants of a gene (2). Mendel’s second principle, the Law of Independent Assortment, states that alleles are assorted independent of each other during gamete formation, leading to individual traits being inherited independently (1). Additionally, Mendel discovered that alleles could either be dominant or recessive. An allele that constituted a phenotypic trait over the other in a heterozygous genotype was labeled dominant, while the other phenotypically unexpressed allele was called recessive (3). A class of diseases was subsequently named after Mendel as they follow the same observations; Mendelian disorders are inherited monogenic diseases that result from mutation at a single gene locus (4). A notable example is phenylketonuria, where loss-of-function mutations in the PAH gene cause systemic excess phenylalanine, resulting in behavioral abnormalities (5).
Mendel’s Laws still provide important insight in understanding Mendelian traits. For example, the Law of Segregation created the basis of dominant and recessive phenotypic ratios (6). The phenotypic ratios in family pedigrees thus allow inference of dominant and recessive traits. This is additionally helpful when an unknown disorder is found to be a Mendelian trait. Since Mendelian traits have complete penetrance, i.e. individuals carrying the pathogenic variant always express the associated trait, it is possible to search for the gene-of-interest when parental genomes are also sequenced. In present-day analysis, Whole Exome Sequencing leverages the fact that most complete penetrance genes lie in the coding region of the genome; this reduces cost and search space for identifying novel diseases (7).
We now know that the Law of Independent Assortment is applicable only when traits are located on different chromosomes. Therefore, it is important in laying the assumption of the lack of linkage between different traits whose loci are genetically far apart. Traditionally, linkage analysis used this prerequisite to identify specific loci within the disease-causing organism, as genes in proximity are often in linkage and do not sort independently (7). Regardless, this stipulation could lead to the belief that the Law of Independent Assortment has less direct value in understanding Mendelian disorders.
In contrast to monogenic diseases, complex diseases arise from multiple genetic and/or environmental factors, displaying complicated inheritance and genetics (1,8). Asthma, for example, was shown to be associated with more than 100 genes with significant inter-population variation (9), and is clinically associated with environmental allergens. Researchers are still looking for contributing variants of many common complex diseases as, unlike Mendelian Disorders, the additive inheritance explained by the associated variants does not explain the genetic contribution to the disease determined by twin studies (8). This is known as the ‘missing heritability problem’, and has prompted scientists to look for other clues.
One way to unravel complex disease genetics lies in the functional characterization of gene variants. Mendelian Diseases thus became an important way to study the link between the genotype-phenotype relationship due to a clear causal relationship and complete penetrance. This puts us in a better position to understand why a variant results in a phenotypic trait (6). Moreover, Mendelian traits allow us to elucidate the functional perturbation due to the mutation itself, providing an excellent opportunity to understand how a change in RNA/protein function caused by mutations can contribute to pathogenesis (6). When variants within complex traits, whether rare or common, are involved within neighboring variants of Mendelian traits, molecular insight may be provided regarding the pathways involved in pathogenesis. Therefore, studying the molecular basis of Mendelian traits could provide essential clues to the bigger puzzle of complex disease.
In the late 2000s, Genome-Wide Associated Studies focused on complex traits and forced Mendelian Diseases to take a back seat; yet today we find that many genetic variants must first be understood through studying Mendelian Diseases. While most Mendelian Diseases are low in incidence, they nonetheless provide valuable lessons as we continue on our journey to understand human genetics.
Citations/References:
- Kennedy, M.A. (2005). Mendelian Genetic Disorders. In eLS, (Ed.). https://doi.org/10.1038/npg.els.0003934
- Cooper, G. M. (2000). The Cell: A Molecular Approach. 2nd edition. NCBI. Retrieved 2022, from https://www.ncbi.nlm.nih.gov/books/NBK9944/
- Wanjin, X., & Morigen, M. (2015). Understanding the cellular and molecular mechanisms of dominant and recessive inheritance in genetics course. Yi chuan = Hereditas, 37(1), 98–108. https://doi.org/10.16288/j.yczz.2015.01.014
- Prosen, T., & Hogge, W. (2008). Molecular and Mendelian Disorders. The Global Library of Women’s Medicine. https://www.glowm.com/section-view/heading/Molecular%20and%20Mendelian%20Disorders/item/223#.YhygEJNBzAN
- MedlinePlus. (2021). Phenylketonuria. https://medlineplus.gov/genetics/condition/phenylketonuria/
- Mendel, G., & Bateson, W. (2013). Mendel’s Principles of Heredity Dover Books on Biology. Courier Corporation.
- Antonarakis, S. E., Chakravarti, A., Cohen, J. C., & Hardy, J. (2010). Mendelian disorders and multifactorial traits: the big divide or one for all?. Nature reviews. Genetics, 11(5), 380–384. https://doi.org/10.1038/nrg2793
- What are complex or multifactorial disorders?: MedlinePlus Genetics. (2021). Medline Plus. https://medlineplus.gov/genetics/understanding/mutationsanddisorders/complexdisorders/
- Allergic asthma: MedlinePlus Genetics. (2020). MedlinePlus. https://medlineplus.gov/genetics/condition/allergic-asthma/
2 nd Place: Gillian Wells, Grade 11 Teacher: Mrs. Rebecca Hodgson School: Ulverston Victoria High School Location: Ulverston, England, UK
Mendel is often referred to as the “Father of Modern Genetics” (1). Prior to his experiments in plant hybridization, it was believed inherited traits resulted from blending the traits of each parent (2). From his studies, Mendel derived three principles of inheritance: the laws of dominance (in a heterozygote, the dominant allele conceals the presence of the recessive allele), segregation (each individual possesses two alleles for a specific trait, one inherited from each parent, and segregated during meiosis) and independent assortment (alleles for separate traits are inherited independently) (3, 4).
These principles give a pattern of inheritance followed by Mendelian or monogenic disorders – disorders caused by variation in a single gene (5). Mendel’s law of dominance explains the pattern of inheritance for autosomal dominant monogenic disorders, which present in individuals with only one dominant mutated allele (2). The heredity of dominant disorders – for example, Huntington’s disease and myotonic dystrophy – therefore follow the same pattern as the dominant traits Mendel observed in pea plants (4, 6). Mendel’s law of dominance also explains the pattern of inheritance for autosomal recessive monogenic disorders, which are not expressed in heterozygous individuals (carriers) as the dominant allele ‘hides’ the mutated recessive allele. Therefore, in families with multiple affected generations, the disorder will appear to ‘skip’ generations, only presenting in individuals that inherit two recessive mutated alleles of the same gene, one from each parent, as explained by Mendel’s law of segregation (2). The heredity of recessive disorders – for example, phenylketonuria and cystic fibrosis – therefore follow the same pattern as the recessive traits Mendel observed in pea plants (4, 6).
This understanding of inheritance patterns establishes the causal relationship between genes and Mendelian disorders, between genotype and phenotype (7). From this, many Mendelian disorder gene identification approaches have been developed, from positional cloning and linkage mapping to whole exome and genome sequencing (8, 9). The results are compiled in Online Mendelian Inheritance in Man (OMIM), a comprehensive database of human genes and genetic disorders, with over 26,000 entries describing over 16,000 genes and 9,000 Mendelian phenotypes (10, 11). Identifying these causal genes improves understanding of specific Mendelian disorders, allowing for molecular diagnosis and carrier testing (9).
In contrast, complex or polygenic diseases are caused by variation in multiple genes interacting with environmental and lifestyle factors, and so do not follow Mendelian inheritance patterns (12). However, widespread comorbidity between Mendelian disorders and complex diseases has been identified, suggesting a genetic association (14). Recent studies have shown that nearly 20% of the identified genes underlying Mendelian disorders contain variants responsible for genome-wide association study (GWAS) signals that cause complex diseases. 15% of all genes underlie Mendelian disorders. Mendelian genes are therefore enriched in GWAS signals and so contribute to the etiology of corresponding complex diseases (13, 14).
Given that different variants of the same gene can give rise to several different phenotypes, some Mendelian genes carry variants that contribute to complex diseases as well as causal variants for Mendelian disorders (13, 15). For example, the gene ABCA4 causes the monogenic conditions retinitis pigmentosa and Stargardt disease, as well as the complex disease age-related macular degeneration (15). Therefore, selecting genes that cause Mendelian disorders for candidate gene association studies can reveal variants that contribute to the etiology of complex diseases, allowing their genetic basis to be understood (10).
Given this genetic association between Mendelian disorders and complex diseases, the identification of Mendelian genes and knowledge of their expression can be used to further understand the mechanisms of associated complex diseases. An example in cardiovascular disease (CVD) research is the identification of causal genes for the monogenic disorder severe hypercholesterolemia. This has provided invaluable insights into lipid transport, leading to an improved understanding of CVD. From this, successful therapies have been developed for CVD using knowledge of the relevant genes and pathways (16). Mutation mechanisms observed in Mendelian disorders that can provide insight into complex disease include anticipation, gene dosage effects, and uniparental disomy (10).
Overall, Mendel’s discoveries revolutionized genetics, creating a model of inheritance that led to advancements in the diagnosis, treatment, and genetic understanding of inherited Mendelian disorders. In turn, research of Mendelian disorders has provided an understanding of the causes and mechanisms of complex diseases through genetic association – up to 23% of genes known to cause Mendelian disorders have been associated with a complex disease (17). The study of Mendelian phenotypes has and will continue to provide breakthroughs in the development of treatments and therapies of all genetic disorders (10).
References/Citations:
- Dastur, AdiE, and PD Tank. “Gregor Johann Mendel: The Father of Modern Genetics.” Journal of Prenatal Diagnosis and Therapy, vol. 1, no. 1, 2010, p. 3, https://doi.org/10.4103/0976-1756.62132.
- Reyna, Barbara, and Rita Pickler. “Patterns of Genetic Inheritance.” Neonatal Network, vol. 18, no. 1, Feb. 1999, pp. 7–10, https://doi.org/10.1891/0730-0832.18.1.7.
- Miko, Ilona. “Gregor Mendel and the Principles of Inheritance | Learn Science at Scitable.” Nature.com, 2014, www.nature.com/scitable/topicpage/gregor-mendel-and-the-principles-of-inheritance-593/#.
- Mendel, Gregor. “Versuche Über Pflanzen-Hybriden.” Der Züchter, vol. 13, no. 10-11, Oct. 1941, pp. 221–68, https://doi.org/10.1007/bf01804628.
- Jensen, Peter K. A. “[Monogenic Hereditary Diseases].” Ugeskrift for Laeger, vol. 165, no. 8, Feb. 2003, pp. 805–9, pubmed.ncbi.nlm.nih.gov/12625123/.
- Chial, Heidi. “Gregor Mendel and Single-Gene Disorders | Learn Science at Scitable.” Nature.com, 2014, www.nature.com/scitable/topicpage/mendelian-genetics-patterns-of-inheritance-and-single-966/.
- Hansen, Adam W., et al. “A Genocentric Approach to Discovery of Mendelian Disorders.” The American Journal of Human Genetics, vol. 105, no. 5, Nov. 2019, pp. 974–86, https://doi.org/10.1016/j.ajhg.2019.09.027.
- Botstein, David, and Neil Risch. “Discovering Genotypes Underlying Human Phenotypes: Past Successes for Mendelian Disease, Future Approaches for Complex Disease.” Nature Genetics, vol. 33, no. S3, Mar. 2003, pp. 228–37, https://doi.org/10.1038/ng1090.
- Gilissen, Christian, et al. “Unlocking Mendelian Disease Using Exome Sequencing.” Genome Biology, vol. 12, no. 9, 2011, p. 228, https://doi.org/10.1186/gb-2011-12-9-228.
- Antonarakis, Stylianos E., and Jacques S. Beckmann. “Mendelian Disorders Deserve More Attention.” Nature Reviews Genetics, vol. 7, no. 4, Mar. 2006, pp. 277–82, https://doi.org/10.1038/nrg1826.
- Hamosh, Ada. “OMIM Entry Statistics.” Omim.org, omim.org/statistics/entry#.
- SCHORK, NICHOLAS J. “Genetics of Complex Disease.” American Journal of Respiratory and Critical Care Medicine, vol. 156, no. 4, Oct. 1997, pp. S103–9, https://doi.org/10.1164/ajrccm.156.4.12-tac-5.
- Blair, David R., et al. “A Nondegenerate Code of Deleterious Variants in Mendelian Loci Contributes to Complex Disease Risk.” Cell, vol. 155, no. 1, Sept. 2013, pp. 70–80, https://doi.org/10.1016/j.cell.2013.08.030.
- Chong, Jessica X., et al. “The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities.” The American Journal of Human Genetics, vol. 97, no. 2, Aug. 2015, pp. 199–215, https://doi.org/10.1016/j.ajhg.2015.06.009.
- Dean, Michael. “Approaches to Identify Genes for Complex Human Diseases: Lessons from Mendelian Disorders.” Human Mutation, vol. 22, no. 4, Aug. 2003, pp. 261–74, https://doi.org/10.1002/humu.10259.
- Kathiresan, Sekar, and Deepak Srivastava. “Genetics of Human Cardiovascular Disease.” Cell, vol. 148, no. 6, Mar. 2012, pp. 1242–57, https://doi.org/10.1016/j.cell.2012.03.001.
- Spataro, Nino, et al. “Properties of Human Disease Genes and the Role of Genes Linked to Mendelian Disorders in Complex Disease Aetiology.” Human Molecular Genetics, vol. 26, no. 3, Feb. 2017, pp. 489–500, https://doi.org/10.1093/hmg/ddw405.
3 rd Place: Yiyang Zhang, Grade 11 Teacher: Dr. Qiongyu Zeng School: Shanghai High School International Division Location: Shanghai, China
Natural populations are characterized by astonishing phenotypic diversity determined by genes and dynamic environmental factors. In 1865, Gregor Mendel showed how traits are passed between generations through his classical pea crosses, giving us the first insight into the heritable basis of phenotypic variation [1]. Mendel’s findings revolutionized the concept of genotype-phenotype relationships and laid the foundation for modern genetics. However, our understanding of the spectrum and continuum between Mendelian and non-Mendelian diseases remains incomplete, and more work is needed to fully unravel the mechanisms underlying human diseases [2].
Mendelian diseases such as sickle cell anemia are characterized by monogenic genetic defects that result in discrete phenotypic differences [3]. Such Mendelian mutations are thought to segregate in predictable patterns, similar to the simple traits Mendel demonstrated in his pea crosses. Indeed, genetic mapping in family-based studies has led to remarkable discoveries of rare chromosomal abnormalities in patients with Mendelian diseases such as Duchenne muscular dystrophy [4]. However, even monogenic diseases follow a Mendelian inheritance pattern only sporadically. For example, in cystic fibrosis (CF), which has nearly 2000 mutant alleles in the primary causative gene Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) and six other loci associated with but not causing the disease, patients exhibit considerable interindividual variability in symptom severity [5, 6]. Thus, there is no pure Mendelian inheritance [7] or, in other words, there are essentially no simple diseases [8].
In Mendelian diseases, mutations in critical genes are usually embryologically lethal, which explains the low prevalence of Mendelian disorders in natural populations [9]. In contrast, common forms of human disease such as diabetes, heart disease, and cancer occur in previously healthy individuals, and instead of dominant disease-causing alleles, many weak genetic factors exert miniscule and accumulative effects on phenotypic outcomes. This multifactorial nature of complex diseases, which are either oligogenic or polygenic [10], means that they do not strictly adhere to Mendelian inheritance patterns in conventional mapping analyses, as segregation of genetic variants in the recombinant offspring of genetically distinct parents can easily hide extreme phenotypes and mask association signals. Therefore, researchers have developed a threshold model that assumes that there is a distribution of susceptibility for a particular trait in the population and that the trait only occurs when a threshold is exceeded [11]. This model could explain ‘all or none’ phenotypes such as cleft palate and why relatives of affected individuals are at higher risk of multifactorial traits such as hypertension or diabetes than the general population [12].
With the advent of genome-wide association studies (GWAS), which use a large sample of unrelated individuals, significant progress has been made in reliably identifying genes that influence the risk of complex diseases [13]. However, even though many thousands of disease susceptibility loci have been characterized, challenges remain, such as the ‘dark matter of inheritance’ that cannot be assigned for most complex traits [14]. Several explanations have been proposed for this, including numerous low-influence variants, rare variants, poorly recognized structural variants, and inadequate estimation of gene-gene and gene-environment interactions [15].
Gene interaction was first demonstrated in retinitis pigmentosa (RP). Since the structural integrity of retinal photoreceptors depends on the functional complexes formed by Retinal Degeneration Slow (RDS) and Rod Outer segment Membrane protein 1 (ROM1), mutations at discrete loci disrupt digenic interactions and produce the same phenotype as alleles of the same locus [16, 17]. This is a perfect example of how pushing the boundaries of Mendelian genetics can help us unravel the true physiological and cellular nature of complex diseases.
In addition to gene-gene interactions, gene-environment interactions also contribute to quantitative traits and trigger the occurrence of complex diseases such as asthma, which are influenced by numerous genetic and nongenetic factors [18]. Environmental factors can also influence traits epigenetically. For example, the more methyl donors such as folic acid or vitamin B12 are present in the diet of young mice, the higher the frequency of methylation at the CpG site of the agouti gene and the darker the coat coloration in adulthood [19, 20].
Our understanding of the causes of disease has evolved from a simplified paradigm of the Mendelian model (one variant-one disease) to a more sophisticated polygenic model. Expanding Mendelian concepts and constructing theoretical models with higher complexity is the first step toward creating a conceptual continuum between Mendelian and non-Mendelian genetic traits. In the long term, genomics and phenomics will continue to be inexhaustible sources of information to elucidate the genetic architecture of both single gene anomalies and complex diseases and to enable more personalized diagnosis and treatment.
- Mendel, J.G., Versuche u ̈ber Pflanzenhybriden Verhandlungen des naturforschenden Vereines in Brünn, Bd. IV für das Jahr, Abhandlungen. 1865: p. 3-47.
- Badano, J.L. and N. Katsanis, Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet, 2002. 3(10): p. 779-89.
- Steinberg, M.H. and A.H. Adewoye, Modifier genes and sickle cell anemia. Curr Opin Hematol, 2006. 13(3): p. 131-6.
- Monaco, A.P., et al., Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature, 1986. 323(6089): p. 646-50.
- Drumm, M.L., A.G. Ziady, and P.B. Davis, Genetic variation and clinical heterogeneity in cystic fibrosis. Annu Rev Pathol, 2012. 7: p. 267-82.
- Emond, M.J., et al., Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet, 2012. 44(8): p. 886-9.
- Van Heyningen, V. and P.L. Yeyati, Mechanisms of non-Mendelian inheritance in genetic disease. Hum Mol Genet, 2004. 13 Spec No 2: p. R225-33.
- Dean, M., Approaches to identify genes for complex human diseases: lessons from Mendelian disorders. Hum Mutat, 2003. 22(4): p. 261-74.
- Quintana-Murci, L. and L.B. Barreiro, The role played by natural selection on Mendelian traits in humans. Ann N Y Acad Sci, 2010. 1214: p. 1-17.
- Assimes, T.L. and P.S. de Vries, Making the Most out of Mendel’s Laws in Complex Coronary Artery Disease. J Am Coll Cardiol, 2018. 72(3): p. 311-313.
- Wright, S., An Analysis of Variability in Number of Digits in an Inbred Strain of Guinea Pigs. Genetics, 1934. 19(6): p. 506-36.
- Korf, B.R., Basic genetics. Prim Care, 2004. 31(3): p. 461-78, vii.
- Crawford, N.P., Deciphering the Dark Matter of Complex Genetic Inheritance. Cell Syst, 2016. 2(3): p. 144-6.
- Kere, J., Genetics of complex disorders. Biochem Biophys Res Commun, 2010. 396(1): p. 143-6.
- Zuk, O., et al., The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A, 2012. 109(4): p. 1193-8.
- Clarke, G., et al., Rom-1 is required for rod photoreceptor viability and the regulation of disk morphogenesis. Nat Genet, 2000. 25(1): p. 67-73.
- Travis, G.H., et al., Identification of a photoreceptor-specific mRNA encoded by the gene responsible for retinal degeneration slow (rds). Nature, 1989. 338(6210): p. 70-3.
- Papi, A., et al., Asthma. Lancet, 2018. 391(10122): p. 783-800.
- Wolff, G.L., et al., Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J, 1998. 12(11): p. 949-57.
- Waterland, R.A. and R.L. Jirtle, Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol, 2003. 23(15): p. 5293-300.
Honorable Mentions
Lena Chae Glenbrook South High School Glenview, Illinois Teacher: Mrs. Marianne Gudmundsson
Angelina Jolie, a famous actress, underwent bilateral mastectomy and oophorectomy to prevent hereditary breast and ovarian cancer that is prevalent in her family [1]. This was only possible because she was able to predict her risk of developing these cancers in her lifetime, which was substantially high enough to warrant prevention surgery. We now know that germline mutations found in BRCA1/2 genes are responsible for hereditary breast and ovarian cancer syndrome transmitted in an autosomal dominant fashion [2]. This discovery was made possible through progress in genetics which began with Mendel’s experiments in the 1800s [3].
Mendel’s discovery helped us better understand Mendelian disorders that involve single-gene mutations. First, the principles of inheritance found in plants opened up opportunities for scientists to apply their observations to patterns they noticed across human generations. This progress towards human studies from plants, helped scientists dissect human diseases that are inherited in a systematic manner. Second, Mendel’s discoveries allowed us to discover and understand the genetic material known as DNA. Because of Mendel’s observations, Watson and Crick were able to demonstrate the structure of the DNA molecule through their discovery of the double helix [4]. The Human Genome Project led by Craig Venter and Francis Collins laid the foundation for us to locate genes responsible for pathogenesis [5]. Third, understanding both the inheritance pattern of specific human hereditary diseases, along with the knowledge of the sequences in the human genome, contributed to the specific discovery of the mutations in such hereditary diseases. For instance, mutations in the HTT gene can cause Huntington’s disease [6], while mutations in the CFTR gene can cause cystic fibrosis [7]. Due to Mendel’s original discovery and experiments, scientists have been able to link genetics to human pathology.
The study of Mendelian disorders aided in a better understanding of complex diseases in two different ways. First, pedigree studies, or family tree analysis, were used to study monogenic Mendelian disorders with high penetrance; this led to a realization that many human diseases cannot be explained by the Mendelian principle of inheritance. Except for a few hereditary diseases, most human diseases involve more than one gene abnormality when comparing the affected versus unaffected members within a family. This finding led to the concept of stepwise multigene abnormalities and environmental interaction with respect to pathogenesis. Second, Genome-Wide Association Studies (GWAS), which is the population-level study of genes and human diseases, could be understood as an aggregate of linkage analyses based on Mendelian principles [8]. It also extended the field of genetics. GWAS made it possible for scientists to define the role of single DNA mutations in complex diseases. Hundreds of thousands of single-nucleotide polymorphisms (SNPs) can be tested to explore the associations between these variants and disease in larger populations. For example, through the GWAS study, over 40 loci have been found to be associated with Type 2 Diabetes Mellitus (DM) [9]. Another highly heritable psychiatric disorder, schizophrenia, is linked with 108 genetic loci according to a GWAS consisting of more than 150,000 samples [10]. An improved understanding of comprehensive genomic mutations involved in such complex diseases led to the creation of a risk profile score (RPS), which is currently used to predict the risk of such disease development [11].
However, human diseases can sometimes be more than just changes in DNA. Both pedigree analysis and GWAS assume that hereditary diseases can fully be explained by genetic mutations. But epigenetic changes can be equally or more important [12]. Epigenetic processes such as DNA methylation or histone modifications, triggered by environmental and behavioral changes, may turn the target gene expressions “on” or “off”. Furthermore, protein modification may also play a role in pathogenesis. Therefore, to better understand complex diseases, it is critical to utilize both the study of genetics stemming from Mendel’s discoveries, and the non-genetic processes including epigenetics, transcriptomics, and proteomics [12].
In summary, Mendel’s discovery helped us better understand Mendelian disorders but also more complex diseases. Owing to Mendel’s principles of inheritance, scientists are now equipped with platforms and techniques to analyze both Mendelian disorders and complex diseases. Individualized treatments are now made possible through accurate diagnoses including identification of mutations leading to disease. Just as Angelina Jolie was able to prevent hereditary breast and ovarian cancer through germline DNA profiling, further in-depth DNA screening in a population can lead to a significant reduction in the risk of various hereditary and complex diseases.
- Jolie, A. (2013, May 14). My Medical Choice. New York Times, pp. 25–25.
- Rebbeck, T. R., Friebel, T., Lynch, H. T., Neuhausen, S. L., van ’t Veer, L., Garber, J. E., Evans, G. R., Narod, S. A., Isaacs, C., Matloff, E., Daly, M. B., Olopade, O. I., & Weber, B. L. (2004). Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: THE PROSE Study Group. Journal of Clinical Oncology, 22(6), 1055–1062. https://doi.org/10.1200/jco.2004.04.188
- B. (2021, May 21). Gregor Mendel. Biography. https://www.biography.com/scientist/gregor-mendel
- Pray, L. (2008) Discovery of DNA structure and function: Watson and Crick. Nature Education 1(1):100
- Adams, J. (2008) Sequencing human genome: the contributions of Francis Collins and Craig Venter. Nature Education 1(1):133
- Conneally P. M. (1984). Huntington disease: genetics and epidemiology. American journal of human genetics, 36(3), 506–526.
- Gallati S. (2003). Genetics of cystic fibrosis. Seminars in respiratory and critical care medicine, 24(6), 629–638. https://doi.org/10.1055/s-2004-815659
- Manolio T. A. (2010). Genomewide association studies and assessment of the risk of disease. The New England journal of medicine, 363(2), 166–176. https://doi.org/10.1056/NEJMra0905980
- Hakonarson, H., & Grant, S. F. (2011). Genome-wide association studies (GWAS): impact on elucidating the aetiology of diabetes. Diabetes/metabolism research and reviews, 27(7), 685–696. https://doi.org/10.1002/dmrr.1221
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511(7510), 421–427. https://doi.org/10.1038/nature13595
- Xiao, E., Chen, Q., Goldman, A. L., Tan, H. Y., Healy, K., Zoltick, B., Das, S., Kolachana, B., Callicott, J. H., Dickinson, D., Berman, K. F., Weinberger, D. R., & Mattay, V. S. (2017). Late-Onset Alzheimer’s Disease Polygenic Risk Profile Score Predicts Hippocampal Function. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 2(8), 673–679. https://doi.org/10.1016/j.bpsc.2017.08.004
- Centers for Disease Control and Prevention. (2020, August 3). What is epigenetics? Centers for Disease Control and Prevention. Retrieved March 1, 2022, from https://www.cdc.gov/genomics/disease/epigenetics.htm
Aadit Jain International Academy Bloomfield Hills, Michigan Teacher: Mrs. Suzanne Monck
Nearly two centuries ago, Gregor Mendel launched the scientific community into the vast world of genetics and diseases with his experiments on the common pea plant (1,2). Specifically, his principles have been instrumental in the plethora of discoveries that have been made in Mendelian disorders. With around 400 million people worldwide suffering from one of the 7,000 Mendelian disorders, much research today centers on identifying the genetic causes of these diseases (3). While Mendel was unaware of genes and DNA when he conducted his study (2), his discoveries kickstarted the substantial research that scientists have undertaken on Mendelian disorders.
Mendel’s principles have directly allowed scientists to understand how Mendelian disorders are inherited. For example, his notable discovery that phenotypes of recessive traits can skip generations (2) applies to Mendelian disorders in the case of carriers (4). These are individuals who may not display the disorder phenotype but still carry and can pass on the altered gene (4). Therefore, it is essential to analyze pedigrees of affected families to determine whether the disease-causing gene has a dominant or recessive phenotype. Importantly, this knowledge helps genetics professionals understand the risk that individuals have of passing on a disorder (5). For example, a person who suffers from an autosomal dominant disorder bears a 50% chance of passing the affected gene to each offspring (5). In contrast, two heterozygous parents for an autosomal recessive disorder have a 25% chance of having an offspring affected with the disorder with each pregnancy (5).
Mendel’s principles of uniformity, segregation, and independent assortment demonstrate how genes and alleles are inherited (2). However, subsequent research revealed exceptions such as the sex-linked pattern of inheritance (2,6). Contrary to inheritance of autosomal single-gene diseases, males and females receive a different number of copies of the implicated gene for sex-linked disorders due to their respective pairs of sex chromosomes (1). As a result, sex-linked diseases tend to be prevalent in only one gender (1). For example, Hemophilia A, a blood clotting disorder, typically affects only males because it is an X chromosome-linked recessive disease (1). It is evident that although Mendel’s principles have laid a strong foundation of inheritance patterns, the scientific community’s understanding of Mendelian disorders is greatly enhanced through new research.
Mendel’s discoveries have been fundamental in developing effective methods to test for disorders. With the understanding that the same allele codes for a specific phenotype, researchers have individuals with the same phenotype disorder undergo sequencing in order to identify the defective gene (7). Such was the case in 2010, when scientists discovered that the MLL2 gene was responsible for Kabuki syndrome: 7 out of 10 individuals in the group suffered from a loss of function in that gene (7). Since then, with the Matchmaker Exchange (MME) and the Monarch Initiative, there has been an emphasis on sharing phenotype and genotype data in order to discover new Mendelian disorders (7).
Although complex diseases are influenced by several factors and do not fully follow the inheritance patterns (8), investigating Mendelian disorders can provide insight into the implicated genes and pathways in them. By analyzing data from established databases, genetic researchers found that in fact 54% of Mendelian disease genes play a notable role in complex diseases as well (9). Genes underlying both diseases tend to be associated with more phenotypes and protein interactions, so studying them can be quite useful in understanding Mendelian disorders and consequently complex diseases (9). In some cases, individuals diagnosed with complex diseases have an underlying monogenic condition that is the cause (10). This specifically highlights the significance of research techniques for single-gene disorders to investigations of complex diseases. In the case of hypercholesterolemia, for example, monogenic forms of the disease were used to determine the impact of lipid transport and to identify the involved pathways in the development of this complex disease (10).
Research on Mendelian disorders has helped scientists understand gene function and mechanisms overall. Studying single-gene disorders can further provide insight into the genetic pathways of complex diseases (9). In fact, with genome-wide association studies (GWAS) into single nucleotide polymorphisms (SNP), thousands of genes implicated in complex diseases have been identified (9). Although many details of complex diseases have been established, heritable aspects still remain uncertain (9). Overall, knowledge of Mendel’s principles and Mendelian disorders will be essential in this case and others as research delves further into disease processes.
- Chial, Heidi. “Mendelian Genetics: Patterns of Inheritance and Single-Gene Disorders.” Edited by Terry McGuire. Nature Education, 2008, www.nature.com/scitable/topicpage/mendelian-genetics-patterns-of-inheritance-and-single-966/.
- Miko, Ilona. “Gregor Mendel and the Principles of Inheritance.” Edited by Terry McGuire. Nature Education, 2008, www.nature.com/scitable/topicpage/gregor-mendel-and-the-principles-of-inheritance-593/.
- Ganguly, Prabarna. “NIH funds new effort to discover genetic causes of single-gene disorders.” National Human Genome Research Institute, 15 July 2021, www.genome.gov/news/news-release/NIH-funds-new-effort-to-discover-genetic-causes-of-single-gene-disorders.
- “Carrier.” National Human Genome Institute, www.genome.gov/genetics-glossary/Carrier.
- “If a genetic disorder runs in my family, what are the chances that my children will have the condition?” MedlinePlus, medlineplus.gov/genetics/understanding/inheritance/riskassessment/.
- Nickle, Todd, and Isabelle Barrette-Ng. “3.5: Sex-Linkage- An Exception to Mendel’s First Law.” Biology LibreTexts, 3 Jan. 2021, bio.libretexts.org/Bookshelves/Genetics/Book%3A_Online_Open_Genetics_(Nickle_and_Barrette-Ng)/03%3A_Genetic_Analysis_of_Single_Genes/3.05%3A__Sex-Linkage-_An_Exception_to_Mendels_First_Law.
- Seaby, Eleanor G., et al. “Strategies to Uplift Novel Mendelian Gene Discovery for Improved Clinical Outcomes.” Frontiers in Genetics, 17 June 2021, www.frontiersin.org/articles/10.3389/fgene.2021.674295/full.
- Craig, Johanna. “Complex Diseases: Research and Applications.” Edited by Alexandre Vieira. Nature Education, 2008, www.nature.com/scitable/topicpage/complex-diseases-research-and-applications-748/.
- Jin, Wenfei, et al. “A systematic characterization of genes underlying both complex and Mendelian diseases.” Human Molecular Genetics, vol. 21, no. 7, 20 Dec. 2011, academic.oup.com/hmg/article/21/7/1611/2900796.
- Chong, Jessica X., et al. “The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities.” Science Direct, www.sciencedirect.com/science/article/pii/S0002929715002451.
Sharanya Ravishanker Conestoga High School Berwyn, Pennsylvania Teacher: Mrs. Liz Gallo
Through his genetic experimentations with pea plants, Gregor Mendel established the following Laws of Inheritance that remain critical to our understanding of heredity: The Law of Segregation, The Law of Independent Assortment, and The Law of Dominance (1, 2). In summation, phenotypes—expressed characteristics—are correlated with the type of allele inherited from each parent during gamete formation when genes randomly separate. If an allele is dominant, it is expressed; if an allele is recessive, the associated characteristic will not be displayed unless a matching recessive allele is inherited from the other parent.
These laws and inheritance patterns form the basis of our understanding of Mendelian disorders, rare monogenic diseases caused by alterations—often single-nucleotide polymorphisms (SNPs) and corresponding amino-acid substitutions resulting in the production of unwanted or malfunctioning proteins—in just one of the 25,000 genes in a human genome (3, 4, 5). These mutations typically occur in germline cells, and are thus passed down through DNA to every cell of the offspring (6). Well known Mendelian diseases include cystic fibrosis, sickle cell anemia, and Huntington’s disease.
Through the application of Mendel’s Laws, geneticists have identified five modes of inheritance for Mendelian disorders: autosomal dominant, autosomal recessive, X-linked dominant, X-linked recessive, and mitochondrial (7), paving the way for geneticists to accurately diagnose Mendelian disorders, a step crucial in providing patients with the treatment and specific care they require, as well as revealing significant information vital to the family planning of individuals who carry recessive alleles for threatening disorders. Genealogical records and pedigree analyses have been utilized to trace inheritance through families, but next-generation sequencing technology has gained traction as a method to detect changes in nucleotide orders. Exome-sequencing, for example, focuses on identifying variants in the protein-coding region (exons), and is regarded as cost-effective due to its specificity, focusing on only 1% of the human genome (8, 9, 10). On the other hand, whole genome sequencing can be advocated for due to its capture of DNA variations outside of exons as well as within. Still, as benign polymorphisms are highly prevalent and frequent, entire genome sequencing can make it difficult to prioritize harmful mutations due to the sheer amount of variants shown (9, 11). RNA sequencing can provide support here by quantifying the effect to which a gene is expressed (11, 12 ).
Information gathered from these methods and Mendelian principles regarding dominance also enable geneticists to determine trait-associated gene loci, allowing for a better understanding of protein formation, modification, and function (13). In fact, as Rockefeller University president and accomplished biochemist Dr. Richard Lifton notes, understanding the connection between genes and expressed traits—SNP and product—has served as “starting points for understanding disease and human biology in general”. For example, analysis of a Mendelian form of hypertension resulted in the discovery of a pathway regulating salt reabsorption and potassium secretion in the kidney (14). Similar discoveries of pathways as a result of studies into Mendelian disorders can increase our understanding and ability to treat complex disorders such as cancer, even if these diseases disregard Mendelian principles of inheritance on account of being caused by numerous genetic and environmental factors interacting with one another.
In the same vein, understanding the results of SNP modification allows for research into the genetic susceptibility for various complex disorders and its correlation with environmental exposure. For example, it was determined that individuals whose genotype is homozygous recessive for xeroderma pigmentosum are highly susceptible to UV light related disorders due to mutations in DNA-repairing genes. Similarly, individuals with a mutation in the Alpha-1 gene are at a greater risk for emphysema, especially through smoking, though the mutation itself isn’t causative of the disease (15). The aforementioned linkages between genes and phenotypes would not be possible without the research into Mendelian disorders that revealed crucial information regarding the impacts of individual genes on expressed phenotypes.
Overall, studies into Mendelian diseases—in turn impacted by the understanding of Mendel’s Laws of Inheritance—have contributed significantly to our knowledge of more complex disorders. This knowledge will prove beneficial in developing more efficient medicinal drugs and therapies that effectively target detrimental proteins or alter gene expression to receive desired results (16). As Dr. James Luspki, Professor of Molecular and Human Genetics at Baylor College of Medicine says, “We’re on the threshold of new explanations of disease inheritance and development” (14). Resulting discoveries from studies into Mendelian principles and disorders will undoubtedly clear the way towards greater advancements in our ability to treat complex disorders.
References/Citations: Mendel’s Law of Segregation. 15 Aug. 2020, https://bio.libretexts.org/@go/page/13271. “Inheritance of Traits by Offspring Follows Predictable Rules.” Nature. Scitable by Nature Education, www.nature.com/scitable/topicpage/inheritance-of-traits-by-offspring-follows-predictable-6524925/#:~:text=One%20allele%20for%20every%20gene,same%22)%20for%20that%20allele. Accessed 22 Feb. 2022. Jackson, Maria et al. “The genetic basis of disease.” Essays in biochemistry vol. 62,5 643-723. 2 Dec. 2018, doi:10.1042/EBC20170053 Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: Evolutionary evidence for differences in molecular effects. Paul D. Thomas, Anish Kejariwal. Proceedings of the National Academy of Sciences Oct 2004, 101 (43) 15398-15403; DOI: 10.1073/pnas.0404380101 The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015). https://doi.org/10.1038/nature15393 “Germline Mutation.” National Cancer Institute, www.cancer.gov/publications/ dictionaries/cancer-terms/def/germline-mutation. Accessed 22 Feb. 2022. Genetic Alliance; District of Columbia Department of Health. Understanding Genetics: A District of Columbia Guide for Patients and Health Professionals. Washington (DC): Genetic Alliance; 2010 Feb 17. Appendix B, Classic Mendelian Genetics (Patterns of Inheritance) Available from: https://www.ncbi.nlm.nih.gov/books/NBK132145/ “Exome Sequencing.” Science Direct, 2018, www.sciencedirect.com/topics/ agricultural-and-biological-sciences/exome-sequencing. Accessed 22 Feb. 2022. “What are whole exome sequencing and whole genome sequencing?” MedlinePlus, 28. July 2021, medlineplus.gov/genetics/understanding/testing/sequencing/. Accessed 22 Feb. 2022. Bamshad, M., Ng, S., Bigham, A. et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 12, 745–755 (2011). https://doi.org/10.1038/nrg3031 Byron, S., Van Keuren-Jensen, K., Engelthaler, D. et al. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet 17, 257–271 (2016). https://doi.org/10.1038/nrg.2016.10 Wang, Zhong et al. “RNA-Seq: a revolutionary tool for transcriptomics.” Nature reviews. Genetics vol. 10,1 (2009): 57-63. doi:10.1038/nrg2484 Chial, H. (2008) Rare Genetic Disorders: Learning About Genetic Disease Through Gene Mapping, SNPs, and Microarray Data. Nature Education 1(1):192 Benowitz, Steven. “Centers for Mendelian Genomics uncovering the genomic basis of hundreds of rare conditions.” National Human Genome Research Institute, 6 Aug. 2015, www.genome.gov/news/news-release/ Centers-for-Mendelian-Genomics-uncovering-the-genomic-basis-of-hundreds-of-rare-conditions. Accessed 22 Feb. 2022. Craig, J. (2008) Complex diseases: Research and applications. Nature Education 1(1):184 Heguy, A et al. “Gene expression as a target for new drug discovery.” Gene expression vol. 4,6 (1995): 337-44.
Zhiyuan Shi BASIS International School Hangzhou Hangzhou, China Teacher: Dr. Dongchen Xu
Mendelian theories provided the foundations for the contemporary understanding of heredity. Mendel’s legacy has been particularly beneficial to medical sciences, where research on inheritance patterns of Mendelian disorders has been made possible through utilizing Mendel’s theory. Mendelian theories serve as robust models for evaluating and verifying the inheritance patterns of particular diseases. Even though our current understanding of genetics has moved beyond the Mendelian model, studying certain Mendelian disorders such as oculocutaneous albinism can lead to an improved understanding of complex disorders with polygenic inheritance.
Oculocutaneous albinism is an autosomal-recessive condition caused by the extremely low level of melanin biosynthesis due to mainly four genes (1, 2). Individuals with this illness will also experience whitening of the skin, certain degrees of vision deterioration, and a higher risk of contracting skin cancer due to the lack of dermal melanin (1, 2); understanding the underlying inheritance pattern of albinism would be advantageous towards the prevention of skin cancers. The genetic cause of oculocutaneous albinism can be explained by Mendelian genetics. The disorder is autosomal, meaning neither the gender of the parents nor the gender of the offspring plays a role in its inheritance. The disorder is recessive, meaning both parents must be carriers for birthing an Albino child (3). Through examination of information like the ones above and specific pathology of the disorder, one can establish critical predictions of an offspring’s genotype based on the family’s history. Such analyses enable us to speculate and reconstruct pedigrees for Mendelian disorders using family history. Information regarding Mendelian disorders running in the family and the possible genotypes for offsprings (50% risk of being carriers and 25% risk of being affected) are important to parents seeking family planning suggestions, reinforcing prevention.
Mendelian and non-Mendelian diseases are often regarded as segregated families of genetic disorders. Complex non-Mendelian disorders involve polygenic traits that don’t follow Mendelian disorders’ monogenic properties. However, genes responsible for monogenic diseases correspondingly contribute to the expression of polygenic traits (4). Mendelian disorders are key in providing the individual monogenic components that contribute to complex disease’s polygenic causes. Some of the gene variants responsible for skin pigmentation disorder and skin cancer are the exact genes responsible for the pigment deficiency in the Mendelian disorder oculocutaneous albinism. The 2 most notable ones are variants of the gene TYRP1, a gene coding for the protein tyrosinase-related protein 1, which contributes to melanosome integrity; and gene SLC45A2, which code for a cation exchange protein that transports material required for melanin synthesis into the melanosome (2, 6, 7). Variants of these genes are inherited as monogenic traits, and studies show they contribute to the formation of polygenic skin cancers such as squamous skin cell carcinoma (8). Mendelian inheritance of other variants of the 2 listed genes can even cause other polygenic skin cancers such as melanoma, exhibiting excessive melanin levels. Research showed that heterozygous variants of TYRP1 and SLC45A2 are overrepresented in families with multiple cases of melanoma (9).
Although overrepresentation of SLC45A2 is found in cases of melanoma, variants of the gene can have the opposite effect. A meta-analysis conducted by Ibarrola-Villava et al., 2012, revealed that the SLC45A2 p.Phe374Leu variant had an odds ratio of 0.41 for melanoma (p = 3.50 * 10^-17), enough for concluding that SLC45A2 p.Phe374Leu negatively correlates with melanoma formation (13). This and the previous evidence suggest that factors affecting melanin concentration, one of the key determinants for the presence of different types of polygenic skin cancers, could be partially attributed to the variants of TYRPI and SLC45A2 genes that involve Mendelian inheritance mechanisms.
Another polygenic disorder with Mendelian roots is growth disorder, in which several genes that contribute to the complex disorder of growth disorders are monogenic. For instance, one factor contributing to the common short stature in growth disorders such as dwarfism is the autosomal dominant Mendelian disorder achondroplasia, resulting from the Mendelian inheritance of the mutated FGFR3 gene (10,11). Another monogenic disorder that contributes to growth disorders such as dwarfism is growth hormone deficiency, an autosomal recessive disorder resulting from the mutation and Mendelian inheritance of the mutated GH1 or GHRHR gene (12).
The Mendelian factors underlying both skin cancer and growth disorders demonstrated the value of studying Mendelian inheritance patterns in complex disorders. Although Mendelian diseases only contribute to a small proportion of all known human disorders, understanding their underlying mechanism and pattern, and utilizing them alongside conventional methods for the investigation of complex diseases is of great importance(5), and would produce spectacular innovations in the field of genetics.
- Marçon, C. R., & Maia, M. (2019). Albinism: Epidemiology, genetics, cutaneous characterization, psychosocial factors. Anais brasileiros de dermatologia. Retrieved March 1, 2022, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6857599/
- Grønskov, K., Ek, J., & Brondum-Nielsen, K. (2007, November 2). Oculocutaneous albinism. Orphanet journal of rare diseases. Retrieved March 1, 2022, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2211462/
- Gulani, A. (2021, May 8). Genetics, autosomal recessive. StatPearls [Internet]. Retrieved March 1, 2022, from https://www.ncbi.nlm.nih.gov/books/NBK546620/
- Franić, S., Groen-Blokhuis, M. M., Dolan, C. V., Kattenberg, M. V., Pool, R., Xiao, X., Scheet, P. A., Ehli, E. A., Davies, G. E., van der Sluis, S., Abdellaoui, A., Hansell, N. K., Martin, N. G., Hudziak, J. J., van Beijsterveldt, C. E. M., Swagerman, S. C., Hulshoff Pol, H. E., de Geus, E. J. C., Bartels, M., … Boomsma, D. I. (2015, October). Intelligence: Shared genetic basis between Mendelian disorders and a polygenic trait. European journal of human genetics : EJHG. Retrieved March 1, 2022, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4592100/
- Lango Allen, H., Estrada, K., Lettre, G., Berndt, S. I., Weedon, M. N., Rivadeneira, F., Willer, C. J., Jackson, A. U., Vedantam, S., Raychaudhuri, S., Ferreira, T., Wood, A. R., Weyant, R. J., Segrè, A. V., Speliotes, E. K., Wheeler, E., Soranzo, N., Park, J.-H., Yang, J., … Hirschhorn, J. N. (2010, October 14). Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. Retrieved March 1, 2022, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2955183/
- Del Bino, S., Duval, C., & Bernerd, F. (2018, September 8). Clinical and biological characterization of skin pigmentation diversity and its consequences on UV impact. International journal of molecular sciences. Retrieved March 1, 2022, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6163216/
- Federico, J. R. (2021, August 27). Albinism. StatPearls [Internet]. Retrieved March 1, 2022, from https://www.ncbi.nlm.nih.gov/books/NBK519018/
- Board, P. D. Q. C. G. E. (2009, July 29). Genetics of Skin Cancer (PDQ®). PDQ Cancer Information Summaries [Internet]. Retrieved March 1, 2022, from https://www.ncbi.nlm.nih.gov/books/NBK65895/
- Nathan, V., Johansson, P. A., Palmer, J. M., Howlie, M., Hamilton, H. R., Wadt, K., Jönsson, G., Brooks, K. M., Pritchard, A. L., & Hayward, N. K. (2019). Germline variants in oculocutaneous albinism genes and predisposition to familial cutaneous melanoma. Pigment Cell & Melanoma Research, 32(6), 854–863. https://doi.org/10.1111/pcmr.12804
- Krakow, D., & Rimoin, D. L. (2010, April 27). The skeletal dysplasias. Nature News. Retrieved March 1, 2022, from https://www.nature.com/articles/gim201054
- U.S. National Library of Medicine. (2020, August 18). Achondroplasia: Medlineplus genetics. MedlinePlus. Retrieved March 1, 2022, from https://medlineplus.gov/genetics/condition/achondroplasia/#inheritance
- U.S. National Library of Medicine. (2020, August 18). Isolated growth hormone deficiency: Medlineplus Genetics. MedlinePlus. Retrieved March 1, 2022, from https://medlineplus.gov/genetics/condition/isolated-growth-hormone-deficiency/
- Ibarrola-Villava, M., Hu, H.-H., Guedj, M., Fernandez, L. P., Descamps, V., Basset-Seguin, N., Bagot, M., Benssussan, A., Saiag, P., Fargnoli, M. C., Peris, K., Aviles, J. A., Lluch, A., Ribas, G., & Soufir, N. (2012). MC1R, SLC45A2 and Tyr genetic variants involved in melanoma susceptibility in southern European populations: Results from a meta-analysis. European Journal of Cancer, 48(14), 2183–2191. https://doi.org/10.1016/j.ejca.2012.03.006
Audric Thakur Reading School Reading, United Kingdom Teacher: Ms. Francis Howson
Mendel’s research intended to determine how characteristics of an individual were inherited by their offspring. At the time, the scientific community lacked the genotypic knowledge required to explain how genetic information was transferred to an individual¹. Only in 1826 did Augustin Sageret discover the idea of trait dominance³ (amid a cultural resurgence of Preformation Theory²), and so it was through observational study that Mendel developed the laws of heredity which ground our understanding of Mendelian disorders today.
Most famous of Mendel’s work are those regarding the rugosus locus and the presence or absence of the SBE1 gene⁴, phenotypically expressed by the distinctive ’round’ or ‘wrinkled’ shapes of pea pods respectively⁵. Specifically, he determined the recessive nature of the wrinkled trait through his monohybrid crossing of a uniformly heterozygous generation of pea plants (which themselves were the progeny of a homozygous-dominant and homozygous-recessive cross)⁵. Naturally, this uniform generation of heterozygous peas all possessed the round characteristic. However, Mendel proved that these peas retained their parents’ ‘elementen’⁵ (or more accurately, DNA), since they went on to produce offspring with characteristics from the grandparent generation, evidenced by the 3:1 ratio of round to wrinkled offspring – clear to us now through use of a Punnett square⁶. Of course, these results were the aggregate of a large sample size across several iterations⁵, and therefore incredibly precise (to the point of controversy⁷). As such, they formed the basis for his laws of heredity.
Deriving Mendel’s laws from his work on pea plants is critical to understanding monogenic conditions because their inheritance patterns are often identical⁸, enabling us to make accurate comparisons between the two. This is demonstrated by the Mendelian condition phenylketonuria (PKU)⁹-¹⁰, an autosomal recessive disorder caused by an absent PAH gene at the genetic locus 12q23.2¹⁰.
Citing national Newborn Screening Reports¹¹, 1.7606% of Caucasian-Americans (1996-2000) are heterozygous carriers of PKU. If I apply some simplified mathematics (i.e. ignoring lifestyle factors), the probability of both parents in a Caucasian-American household being carriers of PKU is 0.0310% (0.017606²). Therefore, as per the rules of inheritance followed by Mendel’s pea plants, 0.0077% (0.0310*0.25) of the Caucasian-American population should be expressors of PKU. According to the National Library of Medicine¹¹, the official estimate is 0.0075% – a remarkable example of the accuracy and utility of Mendel’s work, and how understanding and implementing his discoveries has relevant real-world significance, being comparable to large-scale medical statistics to this day.
Unfortunately, it must be noted that Mendelian disorders are an exceptional minority of genetic conditions – the emerging consensus that most exist on a spectrum from Mendelian conditions¹² (high gene penetrance and low gene-environment interaction¹³) to increasingly complex conditions (incomplete or varying gene penetrance and high gene-environment interaction¹³), and that complex disorders are influenced by a multitude of interconnected factors¹⁴. This is why scientists approach complex disorders by assessing risk of onset, rather than applying Mendelian rules of inheritance. Nevertheless, links between the genotypic expression of Mendelian conditions in an individual and the onset of associated complex disorders have been established in the last decade or so of scientific inquiry¹³.
Studies regarding Mendelian comorbidities alongside complex disorders have proved that genetic loci containing causal variants for both Mendelian disorders and complex disease tend to have a greater influence on the onset of a complex disorder compared to genes that pertain to risk factors for only that complex disorder¹³. This means, for an individual afflicted by a series of Mendelian disorders, the probability that they will develop a complex disorder whose determinant genes are simultaneously involved in expressing those Mendelian disorders is significantly higher¹³. For example, an increased risk of schizophrenia is involved with patients who carry genetic variants of Lujan-Fryns and velo-cardio-facial syndromes¹⁷ (clear correlation), and a higher likelihood of developing type-2 diabetes mellitus if the patient suffers from Huntington’s disease, Friedreich’s ataxia and beta-thalassemia¹⁵-¹⁶ (partially supported correlation). This demonstrates that Mendelian-associated genes are certainly influential in determining emergence of a complex disorder. Therefore, understanding inheritance patterns of these Mendelian conditions is essential to create an accurate way of ascertaining the risk of onset for more complex conditions.
Despite the elusive nature of inheritance patterns surrounding several complex disorders, insight can nevertheless be found in studying genes associated with Mendelian conditions. Due to their high penetrance and straightforward inheritance patterns¹³, these monogenic conditions are easy to diagnose and engage in research with, providing a unique foothold to better understand many complex conditions, and allowing us to form more realistic models to predict their onset¹⁸.
Note: citations from sources published prior to 2015 have been used for historical knowledge or to explain/discuss historical scientific experiments only. The exception to this is reference 15.
- Durmaz, A. A., Karaca, E., Demkow, U., Toruner, G., Schoumans, J., & Cogulu, O. (2015). Evolution of genetic techniques: Past, present, and beyond. BioMed research international. Retrieved February 28, 2022, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4385642/
- Maienschein, J. (2005, October 11). Epigenesis and Preformationism. Stanford Encyclopedia of Philosophy. Retrieved February 28, 2022, from https://plato.stanford.edu/entries/epigenesis/#8
- Zirkle, C. (1951, June). Gregor Mendel & his Precursors. Retrieved February 28, 2022, from https://www.mun.ca/biology/scarr/Zirkle_%281951%29_Gregor_Mendel_&_his_Precursors,%20Isis_42,97-104.pdf
- Smith, A., & Martin, C. (2020, December 11). A history of wrinkled-seeded research in PEA. John Innes Centre. Retrieved February 28, 2022, from https://www.jic.ac.uk/advances/a-history-of-wrinkled-seeded-research-in-pea/
- Miko, I. (2008). Gregor Mendel and the Principles of Inheritance. Nature news. Retrieved February 28, 2022, from https://www.nature.com/scitable/topicpage/gregor-mendel-and-the-principles-of-inheritance-593/
- (while the citation doesn’t reference the SBE1 gene in particular, it does discuss other recessive pea plant traits, making it useful nevertheless) LibreTexts, O. S. (2021, September 22). 8.2: Laws of inheritance. Biology LibreTexts. Retrieved February 28, 2022, from https://bio.libretexts.org/Bookshelves/Introductory_and_General_Biology/Book%3A_Concepts_in_Biology_(OpenStax)/08%3A_Patterns_of_Inheritance/8.02%3A_Laws_of_Inheritace
- Radlick, G. (2015, October 9). Beyond mendelfisher – eprints.whiterose.ac.uk. Beyond the “Mendel-Fisher controversy”. Retrieved February 28, 2022, from https://eprints.whiterose.ac.uk/91201/2/BeyondMendelFisher091015%5B1%5D.pdf
- Chial, H. (2008). Mendelian Genetics: Patterns of Inheritance and Single-Gene Disorders. Nature news. Retrieved February 28, 2022, from https://www.nature.com/scitable/topicpage/mendelian-genetics-patterns-of-inheritance-and-single-966/
- NHS. (2019, December 3). Phenylketonuria. NHS choices. Retrieved February 28, 2022, from https://www.nhs.uk/conditions/phenylketonuria/
- Hillert, A., Anikster, Y., Belanger-Quintana, A., Burlina, A., Burton, B. K., Carducci, C., Chiesa, A. E., Christodoulou, J., Đorđević, M., Desviat, L. R., Eliyahu, A., Evers, R. A. F., Fajkusova, L., Feillet, F., Bonfim-Freitas, P. E., Giżewska, M., Gundorova, P., Karall, D., & Blau, N. (2020, July 14). The genetic landscape and epidemiology of phenylketonuria. The American Journal of Human Genetics. Retrieved February 28, 2022, from https://www.sciencedirect.com/science/article/pii/S0002929720301944
- Arbesman, J., Ravichandran, S., Funchain, P., & Thompson, C. L. (2018, July 1). Melanoma cases demonstrate increased carrier frequency of phenylketonuria/hyperphenylalanemia mutations. Pigment cell & melanoma research. Retrieved February 28, 2022, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6013363/
- 12.Freund, M. K., Burch, K. S., Shi, H., Mancuso, N., Kichaev, G., Garske, K. M., Pan, D. Z., Miao, Z., Mohlke, K. L., Laakso, M., Pajukanta, P., Pasaniuc, B., & Arboleda, V. A. (2018, October 4). Phenotype-specific enrichment of mendelian disorder genes near gwas regions across 62 complex traits. The American Journal of Human Genetics. Retrieved February 28, 2022, from https://www.sciencedirect.com/science/article/pii/S0002929718302854
- Spataro, N., Rodríguez, J. A., Navarro, A., & Bosch, E. (2017, February 1). Properties of human disease genes and the role of genes linked to mendelian disorders in complex disease aetiology. Human molecular genetics. Retrieved February 28, 2022, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5409085/
- Yong, S. Y., Raben, T. G., Lello, L., & Hsu, S. D. H. (2020, July 21). Genetic architecture of complex traits and disease risk predictors. Nature News. Retrieved February 28, 2022, from https://www.nature.com/articles/s41598-020-68881-8
- Blair, D. R., Lyttle, C., Mortensen, J., Bearden, C., Jensen, A., Khiabanian, H., Melamed, R., Rabadan, R., Bernsdam, E., Brunak, S., Jensen, L., Nicolae, D., Shah, N., Grossman, R., Cox, N., White, K., & Rzhetsky, A. (2013, September 26). A Nondegenerate Code of Deleterious Variants in Mendelian Loci Contributes to Complex Disease Risk. Define_me. Retrieved February 28, 2022, from https://www.cell.com/fulltext/S0092-8674(13)01024-6
- (not disproving, but cautioning the results of 15) Montojo, M. T., Aganzo, M., & González, N. (2017, September 29). Huntington’s disease and diabetes: Chronological sequence of its association. Journal of Huntington’s disease. Retrieved February 28, 2022, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5676851/
- Rizvi, S., Khan, A. M., Saeed, H., Aribara, A. M., Carrington, A., Griffiths, A., & Mohit, A. (2018, August 14). Schizophrenia in digeorge syndrome: A unique case report. Cureus. Retrieved February 28, 2022, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6188160/
- Jordan, D., & Do, R. (2018, April 11). Using full genomic information to predict disease: Breaking down the barriers between complex and Mendelian Diseases. Annual Reviews. Retrieved February 28, 2022, from https://www.annualreviews.org/doi/10.1146/annurev-genom-083117-021136
Emma Tse Cheltenham Ladies’ College Cheltenham, United Kingdom Teacher: Ms. Helen Stuart
Between 1856 and 1865, Gregor Mendel conducted experiments on garden peas to investigate inheritance (1). His observations, notably his three principles of inheritance, form the basis of scientists’ grasp of monogenic (Mendelian) disorders today, which are caused by mutations in a single gene (2). Before Mendel’s discoveries, it was widely accepted that traits of progeny were a combination of those of each parent. However, when he cross-pollinated smooth-seeded peas with wrinkled-seeded peas, the offspring (F1 generation) only had smooth seeds as opposed to semi-wrinkled seeds. This gave rise to the concept of dominant traits, as well as his first principle: the principle of uniformity, which states that all offspring of parents with two distinct traits will inherit the same (dominant) trait of one parent (3). Mendel discovered recessive traits by self-pollinating a plant from the F1 generation, noting that its offspring (F2 generation) displayed a 3:1 ratio of smooth to wrinkled seeds (3). This proportion indicated that there was a hidden form of the trait, which Mendel acknowledged passed down to the F2 generation. Mendel also proposed the idea of each parent giving their offspring one heritable unit which he called “elementen”, and scientists now recognise this as genes – more specifically, alleles (2). Sickle-cell anaemia is a well-characterised autosomal recessive disease; those affected inherit two copies of a mutant beta-globin gene (1). Huntington’s disease, on the other hand, is an autosomal dominant disorder in which affected individuals possess at least one copy of the mutant HTT gene (1).
Mendelian disorders are relatively uncommon; on the other hand, complex diseases such as asthma and multiple sclerosis are more prevalent and arise from a combination of genetic, environmental and lifestyle factors (4). Therefore, complex diseases do not entirely adhere to Mendelian inheritance. They can be oligogenic or polygenic, meaning there are multiple genes each with their own mutations contributing to the disease’s phenotype (5). Studying Mendelian disorders allows researchers to examine the mutant gene’s effects on human biochemistry and physiology, thus furthering our understanding of the aetiology of complex, multifactorial diseases (4). An example is obesity, an increasingly pressing medical issue in developed countries. In congenital leptin disorder, a rare disease exhibiting an autosomal recessive inheritance pattern, severe obesity is a typical clinical feature. Affected individuals are unable to produce leptin because of mutations in the leptin encoding gene. Leptin acts on the hypothalamus to halt the production of neuropeptide Y, a neurotransmitter responsible for stimulating food, specifically carbohydrate, intake (6). Thus, studying congenital leptin disorder and other related Mendelian obesity disorders has helped scientists gain deeper insight into the complexity of the underlying causes behind obesity, one of which is the effects of leptin on the human body.
Another example is Van der Woude syndrome, an autosomal dominant condition caused by mutations in the IRF6 gene. It is characterised by a cleft lip and palate, hypodontia and lower lip pits (7). Interestingly, IRF6 mutations were also shown to be associated with non-syndromic isolated cleft lips and palates, which are complex traits and more prevalent in the general population than Van der Woude syndrome (8). This illustrates how the same defective gene could be responsible for rare inherited diseases and common medical conditions simultaneously. In essence, this shows Mendelian disorders and complex diseases that share overlapping phenotypes could be caused by the same sets of genetic aberrations (4).
Furthermore, systematic analyses using statistical methodologies have demonstrated that certain Mendelian disorders and complex diseases share a common genetic foundation. A study examining patients with concomitant Mendelian disorders and cancer revealed genetic connections between the two (9). The researchers’ initial hypothesis was that genetic mutations responsible for certain Mendelian disorders may predispose to the development of cancer. They found that genes associated with melanoma (MC1R and TYR), for instance, are also mutated in patients with oculocutaneous albinism, a Mendelian recessive disorder in which patients lack pigment in their skin, hair or eyes (10). Identifying cancer-driving genes that are found in Mendelian disorders enables scientists to understand the genetic basis of cancer development as well as various clinical presentations in cancer patients.
Although Mendel’s legacy has undoubtedly shaped our present understanding of inheritance, his discoveries alone cannot fully encapsulate the science behind complex diseases. The study of Mendelian disorders has given scientists a strong grounding for further research using advanced technologies such as whole genome sequencing and genome-wide association studies (11, 12), enhancing our knowledge of the genetic mechanisms and pathogenesis underlying polygenic diseases which would have been impossible in the 19th century.
- Molnar, Charles. Concepts Of Biology – 1st Canadian Edition. 1st ed., 2019, pp. Chapter 8.1.
- Chial, Heidi. “Gregor Mendel And Single-Gene Disorders | Learn Science At Scitable”. Scitable By Nature Education, 2008, https://www.nature.com/scitable/topicpage/mendelian-genetics-patterns-of-inheritance-and-single-966/.
- Miko, Ilona. “Gregor Mendel And The Principles Of Inheritance”. Scitable By Nature Education, 2008, https://p75fz1.nbcnews.top/scitable/topicpage/gregor-mendel-and-the-principles-of-inheritance-593.
- Reid, Jeremy. “Rare Disease Research Helps Us Understand Medicine For All Diseases – On Biology”. Biomed Central, 2016, https://blogs.biomedcentral.com/on-biology/2016/02/26/rare-disease-research-helps-understand-medicine-diseases/.
- Collins, Samuel et al. The Genetics Of Allergic Disease And Asthma. 4th ed., Elsevier, 2016, pp. 18-30, https://www.sciencedirect.com/science/article/pii/B9780323298759000033, Accessed 25 Feb 2022.
- Beck, B. “Neuropeptide Y In Normal Eating And In Genetic And Dietary-Induced Obesity”. Philosophical Transactions Of The Royal Society B: Biological Sciences, vol 361, no. 1471, 2006, pp. 1159-1185. The Royal Society, https://doi.org/10.1098/rstb.2006.1855. Accessed 25 Feb 2022.
- Chial, Heidi. “Human Genetic Disorders: Studying Single-Gene (Mendelian) Diseases | Learn Science At Scitable”. Nature.Com, 2008, https://www.nature.com/scitable/topicpage/rare-genetic-disorders-learning-about-genetic-disease-979/.
- Craig, Johanna. “Complex Diseases: Research And Applications”. Nature.Com, 2008, https://www.nature.com/scitable/topicpage/complex-diseases-research-and-applications-748/#:~:text=To%20comprehend%20the%20intricacies%20of,passed%20from%20generation%20to%20generation.
- Melamed, Rachel D. et al. “Genetic Similarity Between Cancers And Comorbid Mendelian Diseases Identifies Candidate Driver Genes”. Nature Communications, vol 6, no. 1, 2015. Springer Science And Business Media LLC, https://doi.org/10.1038/ncomms8033. Accessed 26 Feb 2022.
- “Oculocutaneous Albinism – NORD (National Organization For Rare Disorders)”. NORD (National Organization For Rare Disorders), https://rarediseases.org/rare-diseases/oculocutaneous-albinism/.
- “Genome-Wide Association Studies Fact Sheet”. National Human Genome Research Institute, 2020, https://www.genome.gov/about-genomics/fact-sheets/Genome-Wide-Association-Studies-Fact-Sheet.
- Benowitz, Steven. “Centers For Mendelian Genomics Uncovering The Genomic Basis Of Hundreds Of Rare Conditions”. National Human Genome Research Institute, 2015, https://www.genome.gov/news/news-release/Centers-for-Mendelian-Genomics-uncovering-the-genomic-basis-of-hundreds-of-rare-conditions.
Hannah Wilson Raphael House Rudolf Steiner School Lower Hutt, New Zealand Teacher: Ms. Sarah McKenzie
From his study of pea plants, Gregor Mendel developed three fundamental principles of inheritance: the principle of uniformity, the principle of segregation, and the principle of independent assortment (1). All monogenic traits follow these principles and are thus called Mendelian traits (1,2). Therefore, Mendel’s principles can be used to study Mendelian diseases, notably through pedigree analysis (1,2). The study of Mendelian diseases can in turn provide valuable insight into complex (non-Mendelian) diseases due to genetic correlations between Mendelian and complex diseases (3-6).
Mendel’s principles enable us to both decipher the past inheritance and predict the future inheritance of Mendelian diseases through pedigree analysis. Pedigree charts are diagrams based on Mendel’s principles that visually represent a family’s inheritance history of a Mendelian trait (1,2). Analysis of pedigree charts reveals whether the allele responsible is dominant or recessive, autosomal or sex-linked, due to the specific inheritance pattern exhibited by each allele type (2). Autosomal recessive diseases such as phenylketonuria (PKU) and sickle cell anemia can skip generations because two heterozygous (carrier) parents can give rise to progeny with either the affected or wild-type phenotype (2). Autosomal dominant diseases never skip generations unless random mutation occurs (2). Conversely, sex-linked Mendelian diseases display unique inheritance patterns depending on whether the disease is X-linked or Y-linked, dominant or recessive (2).
Pedigree analysis is applied in genetic counselling (7). Genetic counsellors presented with the family history of two individuals can predict the probability of each possible genotype and phenotype occurring in future offspring (7). These probabilities equip individuals with the information they need to make an informed reproductive decision. Furthermore, the simplicity of Mendel’s principles makes them accessible to the general public, better enabling individuals to understand the nature of their or their loved one’s disease. Nowadays, fetuses can be screened for common genetic defects during pregnancy, however, pedigree analysis maintains its value in that it can provide preliminary information before conception (8).
Although Mendel’s principles form the foundation of inheritance, most human diseases are complex, meaning they violate Mendel’s principles of inheritance (3). Examples of complex diseases include schizophrenia, hypertension, multiple sclerosis, and Alzheimer’s disease (3). Complex diseases are polygenic, meaning they are influenced by multiple genes, and are subject to environmental influence (3). Some also exhibit pleiotropy and epistatic interactions (9,10). Thus, unlike Mendelian diseases, complex diseases lack distinct inheritance patterns (3,4). This poses a challenge to geneticists when attempting to predict an individual’s risk of developing a complex disease.
In addition, there is now evidence that Mendelian and complex diseases are more interconnected than scientists formerly believed (11). For example, cystic fibrosis, typically categorized as an autosomal recessive Mendelian disease, is now believed to involve multiple loci (5,6). A mutation in the CTFR gene, which codes for a membrane channel protein for chlorine ions, forms the primary genetic basis for cystic fibrosis (6,12). However, variation in the severity of cystic fibrosis has been linked to potential modifier genes separate from the CTFR gene (5,6). As eukaryotic gene expression involves transcription factors as well as the structural gene(s) underlying a trait, it is highly likely that other Mendelian diseases also have complex aspects (13).
The study of Mendelian diseases can directly inform the study of complex diseases when a Mendelian disease acts as a model for a complex disease. Such is the case for Van der Woude syndrome, a rare autosomal dominant Mendelian disorder caused by mutations in the IRF6 gene (3,14). Symptoms of Van der Woude syndrome include cleft lip, a birth defect where the tissue in the lip does not join up completely before birth (3,14). Statistical studies provide evidence that one of the genes responsible for isolated cleft lip, a complex disorder, is IRF6, the same gene underlying Van der Woude syndrome (3). The discovery of links between other phenotypically similar Mendelian and complex diseases would be highly beneficial when considering that complex diseases are simultaneously challenging to study in isolation and highly prevalent in the general population (3,4).
Mendel’s abstract but fundamental principles of inheritance have paved the way for modern genetics. These principles directly enable both scientists and the general public to comprehend the inheritance of Mendelian diseases (1). The study of Mendelian diseases can also inform our understanding of complex diseases, especially in cases where a complex disease shares an element of its genetic basis with a Mendelian disease (ref). Therefore, despite their rarity, humankind as a whole is certain to benefit from the continued study of Mendelian diseases.
- Miko, I. (2008). Gregor Mendel and the Principles of Inheritance. Nature Education. https://www.nature.com/scitable/topicpage/gregor-mendel-and-the-principles-of-inheritance-593/
- Chial, H. (2008). Mendelian Genetics: Patterns of Inheritance and Single-Gene Disorders. Nature Education. https://www.nature.com/scitable/topicpage/mendelian-genetics-patterns-of-inheritance-and-single-966/
- Craig, J. (2008). Complex Diseases: Research and Applications. Nature Education. https://www.nature.com/scitable/topicpage/complex-diseases-research-and-applications-748/
- MedlinePlus. (2021, May 14). What are complex or multifactorial disorders? https://medlineplus.gov/genetics/understanding/mutationsanddisorders/complexdisorders
- O’Neal, W. K., & Knowles, M. R. (2018). Cystic Fibrosis Disease Modifiers: Complex Genetics Defines the Phenotypic Diversity in a Monogenic Disease. Annual review of genomics and human genetics, 19, 201–222. https://doi.org/10.1146/annurev-genom-083117-021329
- Buschman, H. (2019, December 10). Modifier Gene May Explain Why Some with Cystic Fibrosis are Less Prone to Infection. UC San Diego Health. https://health.ucsd.edu/news/releases/Pages/2019-12-10-modifier-gene-may-explain-why-some-with-cystic-fibrosis-less-prone-to-infection.aspx
- NBIAcure. (2014). Genetic Counselling. http://nbiacure.org/learn/genetic-counseling/
- MedlinePlus. (2021, September 29). Prenatal Testing. https://medlineplus.gov/prenataltesting.html
- Nagel R. L. (2005). Epistasis and the genetics of human diseases. Comptes rendus biologies, 328(7), 606–615. https://doi.org/10.1016/j.crvi.2005.05.003
- Gratten, J. & Visscher, P.M. (2016). Genetic pleiotropy in complex traits and diseases: implications for genomic medicine. Genome Med 8, 78. https://doi.org/10.1186/s13073-016-0332-x
- Jin, W et al. (2012, April 1). A systematic characterization of genes underlying both complex and Mendelian diseases. Human Molecular Genetics, Volume 21, Issue 7, Pages 1611–1624. https://doi.org/10.1093/hmg/ddr599
- MedlinePlus. (2021, July 6). Cystic Fibrosis. https://medlineplus.gov/genetics/condition/cystic-fibrosis/#causes
- Urry, Meyers, N., Cain, M. L., Wasserman, S. A., Minorsky, P. V., & Reece, J. B. (2018). Campbell Biology: Australian and New Zealand Version (11th edition. Australian and New Zealand version.). Pearson Australia.
- Children’s Hospital of Philadelphia. (2022). Van der Woude Syndrome. https://www.chop.edu/conditions-diseases/van-der-woude-syndrome
Emma Youngblood St. John Paul the Great Catholic High School Dumfries, Virginia Teacher: Dr. Clare Kuisell
“I am convinced that it will not be long before the whole world acknowledges the results of my work.” Gregor Mendel published the results of his pea plant experiments in 1865, but it wasn’t until the 1900s that people began to rediscover his work, and even then, it was controversial (Williams & Rudge, 2015). Now, nearly 200 years later, he is known as the father of the science of genetics, and students throughout the world learn about the laws of segregation and independent assortment which originated from Mendel’s observations. Mendel’s discoveries allow us to understand Mendelian disorders because they have been used to identify patterns of inheritance, which can be applied to genes that are known to have influence in complex diseases.
Single gene diseases are often referred to as Mendelian diseases–or disorders–and may be inherited in one of several patterns (Genetic Alliance, 2010). An example of such a disease is Marfan syndrome. With an incidence of approximately 1 in 5000 individuals, Marfan syndrome is an autosomal dominant disease that affects the body’s connective tissue (Coelho & Almeida, 2020). Using Mendel’s law of dominance and uniformity, which differentiates dominant and recessive alleles (Lewis & Simpson, 2021), one can predict the inheritance pattern of Marfan syndrome using the same calculations and ratios Mendel discovered in his pea plants. Because the mutated allele of the gene is dominant, a child who inherits Marfan syndrome must have a parent who also has it. This also means that Marfan syndrome, like other autosomal dominant diseases, would occur in every generation until the dominant allele is not inherited from either the mother or the father. Mendel’s work has allowed the identification of different types of inheritance patterns of single gene disorders to be very simple.
Complex diseases, while much less predictable than Mendelian disorders, are still influenced by genetics. Almost all complex diseases are affected by multiple genes and environmental factors, and examples include heart disease, cancer, and diabetes (National Human Genome Research Institute, 2013). Another well-known complex disease is Alzheimer’s Diseases (AD). Approximately 44 million people currently live with AD, and that number is expected to triple by 2050 (Lane et al., 2018). Aside from age, one of the highest risk factors for AD is the presence of the ε4 allele of the gene that codes for apolipoprotein E, also called ApoE (Yin & Wang, 2018). Recent studies have also shown that two of the most reliable biomarkers for AD are Aβ protein deposits and phosphorylated tau proteins (Mantzavinos & Alexiou, 2017). By studying the genes that code for these proteins and the gene that codes for, scientists may be able to identify a better way to treat or even cure AD. The multiple factors that affect complex diseases make it nearly impossible to determine exact patterns of inheritance, but if single genes that influence them can be isolated, the same patterns used to predict inheritance patterns in Mendelian disorders can be used to predict a high or low likelihood of developing or inheriting a complex disease.
Mendel’s discoveries have been essential in determining the inheritance patterns of Mendelian disorders, which can also be used to form a more accurate prediction of the inheritance of complex diseases. Interest in genetics-related careers is rapidly growing; the U.S. Bureau of Labor Statistics shows a job outlook of 26% from 2020 to 2030. This compares to the outlook of 14% for other healthcare occupations and 8% for all occupations (2021). Increased interest in the field of genetics may lead to new ways of applying the discoveries Mendel made nearly 200 years ago to solve modern questions and problems. It might have taken longer for the world to acknowledge the results of his work than he believed it would, but there is no doubt that once it did, Gregor Mendel’s work opened a realm of new scientific possibilities that will certainly endure for 200 years more.
Boyle, E. A., Li, Y. I., & Pritchard, J. K. (2017). An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell, 169(7), 1177–1186. https://doi.org/10.1016/j.cell.2017.05.038 Coelho, S. G., & Almeida, A. G. (2020). Marfan syndrome revisited: From genetics to the clinic. Síndrome de Marfan revisitada – da genética à clínica. Revista portuguesa de cardiologia, 39(4), 215–226. https://doi.org/10.1016/j.repc.2019.09.008 Genetic Alliance. (2010, February 17). Classic Mendelian Genetics (Patterns of Inheritance). Understanding Genetics: A District of Columbia Guide for Patients and Health Professionals. Retrieved January 20, 2022, from https://www.ncbi.nlm.nih.gov/books/NBK132145/ Lane, C. A., Hardy, J., & Schott, J. M. (2018). Alzheimer’s disease. European journal of neurology, 25(1), 59–70. https://doi.org/10.1111/ene.13439 Lewis, R. G., & Simpson, B. (2021). Genetics, Autosomal Dominant. In StatPearls. StatPearls Publishing. Mantzavinos, V., & Alexiou, A. (2017). Biomarkers for Alzheimer’s Disease Diagnosis. Current Alzheimer research, 14(11), 1149–1154. https://doi.org/10.2174/1567205014666170203125942 National Human Genome Research Institute. (2013, May 3). Genetic Analysis Tools Help Define Nature and Nurture in Complex Disorders. Genome.gov. Retrieved January 20, 2022, from https://www.genome.gov/10000865/complex-disorders-background U.S. Bureau of Labor Statistics. (2021, September 8). Genetic counselors : Occupational outlook handbook. U.S. Bureau of Labor Statistics. Retrieved January 21, 2022, from https://www.bls.gov/ooh/healthcare/genetic-counselors.htm Williams, C. T., & Rudge, D. W. (2015). Mendel and the Nature of Science. The American Biology Teacher, 77(7), 492–499. https://doi.org/10.1525/abt.2015.77.7.3 Yin, Y., & Wang, Z. (2018). ApoE and Neurodegenerative Diseases in Aging. Advances in experimental medicine and biology, 1086, 77–92. https://doi.org/10.1007/978-981-13-1117-8_5
Vivian Yuan Ridgewood High School Ridgewood, New Jersey Teacher: Mr. Ryan Van Treuren
Complex Diseases Through the Lens of Mendelian Genetics
In 2001, the Human Genome Project reported that the human genome contains 20,000 to 25,000 protein-coding genes (1, 2). Among those genes, less than 10% are related to single gene diseases, also known as monogenic or Mendelian disorders (2). With the recent advances of genome-wide association studies (GWAS) and single nucleotide polymorphism (SNP) sequencing approaches, interest in human genetics has shifted from rare Mendelian disorders to more common complex diseases, which involve both genetic components and environmental factors (2, 3, 4). Although Mendelian disorders affect a small portion of the population, studying them has contributed greatly to our understanding of genetic mutations and the risk factors underlying the aetiology of complex diseases.
The foundation of all modern human genetic studies relies upon Gregor Mendel’s study with pea plants. Through his experiments, Mendel discovered three laws: the law of dominance, the law of segregation, and the law of independent assortment (5, 6). Mendelian laws aptly dictate Mendelian disorders, which allows scientists to better determine the inheritance pattern of diseases. Disease inheritance genes can be classified as autosomal or sex linked, dominant or recessive. Huntington’s disease, a progressive neurodegenerative disorder, is an example of autosomal dominant Mendelian disorder, because only one copy of the defective gene from one parent is needed for disease manifestation. Conversely, phenylketonuria (PKU), which causes the accumulation of the amino acid phenylalanine, is an autosomal recessive disease. Both parents must give the defective gene to the child for the disease to appear. If only one parent carries the mutated gene, the child will not be affected, but they could still be a carrier of the mutated gene. Luckily, doctors are now able to predict the genotype and phenotype of an individual using pedigree analysis. Now, PKU could be confirmed within three days after birth, and PKU babies will be switched to a low protein and phenylalanine diet, preventing cognitive abnormality.
Although complex diseases do not follow Mendelian inheritance, the mechanisms learned from Mendelian diseases can help scientists understand complex diseases (2). Initially, cystic fibrosis was characterized as an autosomal recessive monogenic disease because of the mutations in the Cystic Fibrosis Trans-membrane conductance Regulator (CFTR) gene. However, recent studies showed that not all CFTR mutations produce the same disease, and disease severity is associated with modifier genes (7, 8). The interactions between modifier genes and different CFTR mutations heavily affect the phenotypic complexity and expressivity of CFTR genes. Due to the discovery of these modifier genes, cystic fibrosis is now classified as an oligogenic disease, involving a few genes. In a study of several families with epilepsy, multiple members carrying the same SCN1A gene mutations showed varying phenotypes and disease severity. Like the case in cystic fibrosis, modifier genes were also identified in epilepsy. While they may not be pathogenic, those genes still account for the variability in SCN1A-related phenotype (9).
In addition, study of Mendelian diseases can provide useful information about individual gene’s contribution to the phenotypes in complex diseases. When comparing two databases, Online Mendelian Inheritance in Man database (OMIM) and Genetic Association database (GAD), scientists found that among the 968 Mendelian genes identified, 524 genes are also genetic risk factors for complex diseases (3); hence, those genes are called complex-Mendelian genes (CM genes). CM genes were found to have higher allelic Odds Ratios (ORs) than genes associated only with complex disease, suggesting that CM genes have stronger effects on the complex phenotypes they affect (10).
Furthermore, some complex diseases, such as breast cancer and hypertension, have Mendelian subtypes that clearly display the inheritance patterns typical of monogenic diseases. Hereditary breast cancer, accounting for 5%-10% of all breast cancer, is mainly caused by a mutation in BRCA1 and BRCA2 genes (11). The inheritance of BRCA1 and BRCA2 follows an autosomal dominant pattern, and carriers of those two genes are at higher risk of developing other cancers, especially ovarian cancer. Similarly, scientists have found that some types of hypertension, called monogenic hypertension, are caused by distinct genetic mutations resulting in gain-of-function or loss-of-function in the mineralocorticoid, glucocorticoid, or sympathetic pathways (12).
The knowledge gained from studying genetic inheritance is surely invaluable to understanding diseases and finding treatments. Future applications of these basic principles laid out by Mendel over 150 years ago will lead doctors to predict disease manifestation and severity, working towards prevention and early treatment for all diseases, simple or complex.
- International Human Genome Sequencing Consortium (2004) Finishing the Euchromatic Sequence of the Human Genome. Nature 431: 931-945
- Antonarakis S.E. and Beckman J.S. (2006) Mendelian disorders deserve more attention. Nature Reviews Genetics 7: 277-282
- Jin WF, Qin PF, Lou HY and Xu SF. (2012) A systematic characterization of genes underlying both complex and Mendelian diseases. Human Molecular Genetics 21 (7): 1611-1624
- Craig J. (2018) Complex Diseases: Research and Applications. Nature Education 1 (1): 184 https://www.nature.com/scitable/topicpage/complex-diseases-research-and-applications-748/
- Miko I. (2008) Gregor Mendel and the Principles of Inheritance. Nature Education 1 (1): 134. https://www.nature.com/scitable/topicpage/gregor-mendel-and-the-principles-of-inheritance-593/
- Chial H. (2008) Mendelian Genetics: Patterns of Inheritance and Single-Gene Disorders. Nature Education 1 (1): 63. https://www.nature.com/scitable/topicpage/mendelian-genetics-patterns-of-inheritance-and-single-966/
- Buratti E., Brindisi A., Pagani,F. & Baralle F. E. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am. J. Hum. Genet. 74, 1322–1325 (2004).
- O’Neal W.K. and Knowles M.R. Cystic Fibrosis Disease Modifiers: Complex Genetics Defines the Phenotypic Diversity in a Monogenic Disease. Annu. Rev. Genom. Hum. Genet. 2018. 19:201–22
- de Lange I.M., Mulder F., Slot R, et al (2020). Modifier genes in SCN1A-related epilepsy syndromes. Mol Genet Genomic Med. 8: e1103
- Spataro N., Rodriguez J., Navarro A., Bosch, E. (2017) Properties of Human Disease Genes and the Role of Genes Linked to Mendelian Disorders in Complex Disease Aetiology. Human Molecular Genetics 26 (3): 489-500
- Mehrgou A. and Akouchekian M. (2016) The Importance of BRCA1 and BRCA2 gene mutations in breast cancer development. Med J Islam Repub Iran 30: 369
- Raina R, Krishnappa V, Das A, et al (2019) Overview of Monogenic or Mendelian forms of Hypertension. Frontiers in Pediatrics 7: 263
Xinyi Zhang South Brunswick High School Monmouth Junction, New Jersey Teacher: Ms. Jessica Pagone
Genetic mutations lend each person their individuality, but certain variations can cause adverse health effects. Mendelian, or monogenic, disorders arise from variations in just one of the over 4,000 protein-coding genes that are currently associated with these diseases (2). Using Mendel’s principles to trace the inheritance pattern and phenotypes of a specific genetic mutation forms the basis of studying monogenic disorders. In turn, these findings can elucidate the role of various genetic mutations in diseases with more complex causes (8).
Gregor Mendel’s laws of genetic inheritance establish the framework for Mendelian patterns of inheritance. Given that each parent provides an allele for every gene in their offspring, if one parent has a genetic mutation that may cause a certain monogenic disorder, their offspring may inherit the mutant allele (5). Whether the child will develop the disorder or be a carrier depends on the dominance of the alleles they inherit (11).
Coupling Mendel’s principles with pedigree analysis reveal predictable modes of inheritance that bring light to the genetic nature of Mendelian diseases (5). Consider, for example, the realization of the inheritance pattern of sickle cell disease (SCD). Both parents need to have at least one mutant allele in the hemoglobin beta (HBB) gene to produce offspring with SCD (6). However, if their offspring only has one mutant allele, they will not be afflicted with SCD (6). With these observations, scientists determined that SCD is an autosomal recessive disorder in which it could only develop in people with two mutant alleles of the HBB gene (11). The inheritance pattern of a Mendelian disease would be different in an autosomal dominant disorder, where one mutant allele is enough to cause the disease, or in a sex-linked disorder, where diseases are inherited through the X or Y chromosome (11). Using Mendel’s principles to identify Mendelian inheritance patterns often serves as the first step in assessing disease risk and pinpointing the responsible genotype.
In actuality, Mendelian disorders are much rarer than complex disorders, which are distinguished from monogenic conditions because many genes, environmental interactions, and lifestyle choices all contribute to disease development (8). These variables complicate the determination of inheritance patterns or causative factors of a complex disease.
Despite their inherent differences, some connections have been uncovered between Mendelian and complex diseases. Many monogenic diseases are comorbid with complex ones (4). Furthermore, over 20% of the gene variations that cause Mendelian disorders have been implicated in at least one complex disorder (8). For instance, mutations in the IRF6 gene can lead to Van der Woude syndrome, a rare Mendelian disorder that causes cleft lip, cleft palate, and other facial deformities (10). Intriguingly, IRF6 mutations have also been implicated in complex, isolated forms of cleft lip and palate (12). These overlaps highlight the importance of utilizing Mendelian diseases to understand complex disease etiology.
Techniques such as whole-exome sequencing can link the characteristics of a Mendelian disease with the mutant gene that causes them (9). These findings are recorded in the Online Mendelian Inheritance in Man (OMIM), an accessible catalog of thousands of genotype-phenotype links for monogenic disorders (3). Studying this data has led to the identification of mutations and pathways that play a role in producing similar phenotypes in complex diseases (3,4). To better understand the complexity of essential hypertension, researchers studied many Mendelian disorders that are associated with high blood pressure, such as Liddle’s syndrome (7). Many of these disorders are caused by genetic mutations that alter proteins involved in renal salt balance (7). These studies brought attention to the importance of the kidneys and adrenal glands in regulating blood pressure and revealed the genetic mutations that may be associated with essential hypertension (7). Better knowledge of the molecular pathways behind essential hypertension has opened up new targets in drug development, such as ROMK, a renal potassium channel that is altered by a monogenic disorder known as Bartter syndrome type II (1).
Overall, while insights gleaned from studying Mendelian disorders cannot account for the environmental or lifestyle risks that contribute to complex diseases, they can guide research on pinpointing the pathophysiological processes and susceptibility alleles that bring about complex disorders. Thus, despite the rarity of Mendelian disorders, research on them should not be undercut to prioritize the study of prevalent complex diseases. A more comprehensive understanding of Mendelian disorders allows for more efficient risk assessment, prevention measures, and diagnoses for Mendelian and complex diseases alike, rendering it a valuable tool that should be further explored in the field of medical genetics.
- Abdel-Magid, A. F. (2016, November 22). Potential of renal outer medullary potassium (ROMK) channel as treatments for hypertension and heart failure. American Chemical Society. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5238487/
- Antonarakis, S. E. (2021, June 23). History of the methodology of disease gene identification … Wiley Online Library. Retrieved from https://onlinelibrary.wiley.com/doi/10.1002/ajmg.a.62400
- Brownlee, C. (n.d.). OMIM turns 50: A genetic database’s past, present, and future. Johns Hopkins Medicine. Retrieved from https://www.hopkinsmedicine.org/research/advancements-in-research/fundamentals/in-depth/omim-turns-50-a-genetic-databases-past-present-and-future
- Kumar Freund, M. (2018, October 4). Phenotype-Specific Enrichment of Mendelian Disorder Genes near GWAS Regions across 62 Complex Traits. Cell. Retrieved from https://www.cell.com/ajhg/fulltext/S0002-9297(18)30285-4
- Lewis, R. G. (2021, May 7). Genetics, autosomal dominant. StatPearls [Internet]. Retrieved from https://www.ncbi.nlm.nih.gov/books/NBK557512/
- Mangla, A. (2021, December 19). Sickle cell anemia. StatPearls [Internet]. Retrieved from https://www.ncbi.nlm.nih.gov/books/NBK482164/
- Seidel, E., Scholl, U. I. (2017, November 1). Genetic mechanisms of human hypertension and their implications for blood pressure physiology. Physiological Genomics. Retrieved from https://journals.physiology.org/doi/full/10.1152/physiolgenomics.00032.2017
- Spataro, N., Rodríguez, J. A., Navarro, A., & Bosch, E. (2017, February 1). Properties of human disease genes and the role of genes linked to mendelian disorders in complex disease aetiology. Human molecular genetics. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5409085/
- Suwinski, P., Ong, C. K., Ling, M. H. T., Poh, Y. M., Khan, A. M., & Ong, H. S. (2019, February 12). Advancing personalized medicine through the application of whole exome sequencing and Big Data Analytics. Frontiers. Retrieved from https://www.frontiersin.org/articles/10.3389/fgene.2019.00049/full
- U.S. National Library of Medicine. (2020, August 18). Van der Woude Syndrome: Medlineplus Genetics. MedlinePlus. Retrieved from https://medlineplus.gov/genetics/condition/van-der-woude-syndrome/
- 11. U.S. National Library of Medicine. (2021, April 19). What are the different ways a genetic condition can be inherited?: Medlineplus Genetics. MedlinePlus. Retrieved from https://medlineplus.gov/genetics/understanding/inheritance/inheritancepatterns/
- Zhao, H., Zhang, M., Zhong, W., Zhang, J., Huang, W., Zhang, Y., Li, W., Jia, P., Zhang, T., Liu, Z., Lin, J., & Chen, F. (2018, July 20). A novel IRF6 mutation causing non-syndromic cleft lip with or without cleft palate in a pedigree. OUP Academic. Retrieved from https://academic.oup.com/mutage/article/33/3/195/5056500
ASHG uses cookies to provide you with a secure and custom web experience. Privacy Policy
Home — Essay Samples — Science — Genetic Modification — The Power of Genetic Modification
The Power of Genetic Modification
- Categories: Bioethics Genetic Engineering Genetic Modification
About this sample

Words: 1203 |
Published: Aug 30, 2022
Words: 1203 | Pages: 3 | 7 min read

Cite this Essay
To export a reference to this article please select a referencing style below:
Let us write you an essay from scratch
- 450+ experts on 30 subjects ready to help
- Custom essay delivered in as few as 3 hours
Get high-quality help

Verified writer
- Expert in: Science

+ 120 experts online
By clicking “Check Writers’ Offers”, you agree to our terms of service and privacy policy . We’ll occasionally send you promo and account related email
No need to pay just yet!
Related Essays
2 pages / 948 words
1 pages / 537 words
2 pages / 720 words
1 pages / 471 words
Remember! This is just a sample.
You can get your custom paper by one of our expert writers.
121 writers online
Still can’t find what you need?
Browse our vast selection of original essay samples, each expertly formatted and styled
Related Essays on Genetic Modification
GMO effects on the environment have become a subject of intense debate and scrutiny in recent years. Genetically Modified Organisms (GMOs) have revolutionized agriculture by altering the genetic makeup of crops to enhance their [...]
Genetically Modified Organisms, commonly known as GMOs, have become a controversial topic in today's society. These organisms have been altered through genetic engineering techniques to improve their characteristics or introduce [...]
Genetic editing has been a topic of discussion for decades. With advances in science, genetic editing has become a reality, and many are debating the ethical implications of this technology. While some argue that genetic editing [...]
Functional RNAs are produced through modification of pre-RNA produced from transcription process except bacterial mRNAs which are as such used for protein synthesis without any modification. These series of modification [...]
WAGR Syndrome is an uncommon disease that can affect both genders. It is more common for babies to be born with the syndrome, rather than getting diagnosed with the disease later on in life. The babies that are born with WAGR [...]
It is known that children who have siblings diagnosed with an autism-spectrum disorder have a greater risk of themselves becoming diagnosed with an autism spectrum disorder than do other similar children who do not have a family [...]
Related Topics
By clicking “Send”, you agree to our Terms of service and Privacy statement . We will occasionally send you account related emails.
Where do you want us to send this sample?
By clicking “Continue”, you agree to our terms of service and privacy policy.
Be careful. This essay is not unique
This essay was donated by a student and is likely to have been used and submitted before
Download this Sample
Free samples may contain mistakes and not unique parts
Sorry, we could not paraphrase this essay. Our professional writers can rewrite it and get you a unique paper.
Please check your inbox.
We can write you a custom essay that will follow your exact instructions and meet the deadlines. Let's fix your grades together!
Get Your Personalized Essay in 3 Hours or Less!
We use cookies to personalyze your web-site experience. By continuing we’ll assume you board with our cookie policy .
- Instructions Followed To The Letter
- Deadlines Met At Every Stage
- Unique And Plagiarism Free

COMMENTS
To help you get started, here are 115 genetics essay topic ideas and examples to inspire your writing: The impact of genetic engineering on agriculture. The ethics of genetic manipulation in humans. The role of genetics in determining intelligence. Genetic disorders: causes and treatments. The genetics of cancer.
Genetics is the branch of science concerned with genes, heredity, and variation in living organisms. It seeks to understand the process of trait inheritance from parents to offspring, including ...
Genetics forms one of the central pillars of biology and overlaps with many other areas, such as agriculture, medicine, and biotechnology. Since the dawn of civilization, humankind has recognized the influence of heredity and applied its principles to the improvement of cultivated crops and domestic animals. A Babylonian tablet more than 6,000 ...
This essay delves into the key principles of genetics, exploring its role in heredity, evolution, and the remarkable strides made in biotechnology. Heredity and the Blueprint of Life: At the heart of genetics lies the concept of heredity, the transmission of genetic information from one generation to the next. Genes, the units of heredity, are ...
12. Essay on Mutations: Mutations are errors in the genotype that create new alleles and can result in a variety of genetic disorders. In order for a mutation to be inherited from one generation to another, it must occur in sex cells, such as eggs and sperm, rather than in somatic cells. The best way to detect a genetic disorder is karyotyping.
Genetics. "Half of your DNA is determined by your mother's side, and half is by your father. So, if you seem to look exactly like your mother, perhaps some DNA that codes for your body and how ...
Genetic Variation and Human Evolution. Department of Human Genetics University of Utah School of Medicine. The past two decades have witnessed an explosion of human genetic data. Innumerable DNA sequences and genotypes have been generated, and they have led to significant biomedical advances. In addition, these data have greatly increased our ...
76 essay samples found. Genetics is the study of genes, genetic variation, and heredity in organisms. Essays on genetics might delve into the fundamental principles of genetics, the discovery and function of DNA, or the development of genetic technologies like CRISPR. Other discussions could explore ethical issues related to genetic engineering ...
Genetics is the study of genes and tries to explain what they are and how they work. Genes are how living organisms inherit features or traits from their ancestors; for example, children usually look like their parents because they have inherited their parents' genes. Genetics tries to identify which traits are inherited and to explain how these traits are passed from generation to generation.
Genetics, the science of biological inheritance, has made extraordinary progress since its simple beginnings in the 1850s. It all started in a Czech monastery garden as the lone studies of a monk, Gregor Mendel. Mendel's pivotal work was at the time largely ignored, and was almost lost to science.
In celebration of the 20th anniversary of Nature Reviews Genetics, we asked 12 leading researchers to reflect on the key challenges and opportunities faced by the field of genetics and genomics.
Essays Biochem (2018) 62 (5): 643-723. Genetics plays a role, to a greater or lesser extent, in all diseases. Variations in our DNA and differences in how that DNA functions (alone or in combinations), alongside the environment (which encompasses lifestyle), contribute to disease processes.
Genetics of Bacteria. Classical genetic analyses of bacteria have made numerous and profound contributions to both fundamental and applied science. From early studies on gene regulation and the nature of horizontal gene transfer, through to the recent discovery and harnessing of CRISPR, bacterial genetics continues to yield important advances.
Louisa A. Stark, Ph.D., is director of the Genetic Science Learning Center (GSLC) at the University of Utah. During graduate school, Stark had the opportunity to take hands-on science into Denver inner-city schools as a member of the "Science Squad," sparking a passion for working with kindergarten through grade-12 students and teachers.
ASHG is proud to support National DNA Day through the Annual DNA Day Essay Contest. DNA Day commemorates the completion of the Human Genome Project in April 2003 and the discovery of the double helix of DNA in 1953. This contest is open to students in grades 9-12 worldwide and asks students to examine, question, and reflect on important ...
One of the fundamental areas of this development is genetic research. Thus, the study of genetics makes it possible to solve pressing issues related to medicine, forensics, and security. This essay aims to discuss the public importance of supporting genetic research. Get a custom essay on The Study of Genetics: Importance for Society.
Genetic technologies1—the ability to manipulate and trans- form the properties of cells, seeds, microbes, insects, plants, animals and even humans—are pushing the frontiers of science and ...
April 25, 2022 DNA Day. 1st Place: Man Tak Mindy Shie, Grade 12. Teacher: Dr. Siew Hwey Alice Tan. School: Singapore International School (Hong Kong) Location: Hong Kong, China. Many would say that the most significant stride in recent genetics has been the completion of the human genome, which laid the basis for studying genetic variation.
Absolutely FREE essays on Genetics. All examples of topics, summaries were provided by straight-A students. Get an idea for your paper ... Introduction DNA profiling, also known as genetic fingerprinting, has revolutionized the fields of forensic science, medicine, and genealogy. Developed in the mid-1980s by Dr. Alec Jeffreys, DNA profiling ...
The process of genetic modification is as follows, first researchers create a small piece of RNA with a short "guide" sequence that attaches (binds) to a specific target sequence of DNA in a genome. This RNA also binds to the Cas9 enzyme. The modified RNA is used to recognize the DNA sequence and the Cas9 enzyme cuts the DNA at the targeted ...